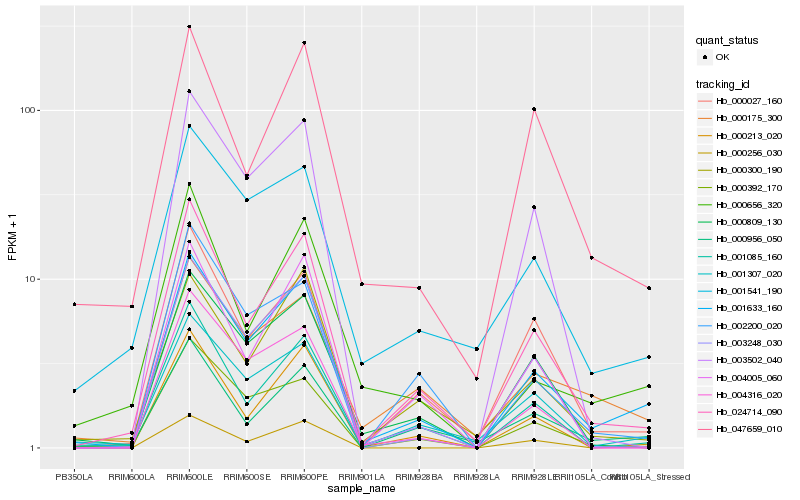
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001633_160 |
0.0 |
- |
- |
PREDICTED: zerumbone synthase-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_024714_090 |
0.1159162054 |
rubber biosynthesis |
Gene Name: SRPP3 |
PREDICTED: stress-related protein [Jatropha curcas] |
| 3 |
Hb_001307_020 |
0.132439395 |
- |
- |
electron carrier, putative [Ricinus communis] |
| 4 |
Hb_001085_160 |
0.1385065356 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 5 |
Hb_000027_160 |
0.1468334463 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 6 |
Hb_000256_030 |
0.1475828117 |
- |
- |
transferase, putative [Ricinus communis] |
| 7 |
Hb_000175_300 |
0.1477250639 |
- |
- |
PREDICTED: uncharacterized protein LOC105634858 [Jatropha curcas] |
| 8 |
Hb_000213_020 |
0.1498851611 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003248_030 |
0.1515041043 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_000956_050 |
0.1527967893 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000809_130 |
0.1554684459 |
- |
- |
PREDICTED: peroxidase 10 isoform X3 [Jatropha curcas] |
| 12 |
Hb_000300_190 |
0.1566915309 |
- |
- |
PREDICTED: uncharacterized protein LOC105632566 [Jatropha curcas] |
| 13 |
Hb_000392_170 |
0.1583262956 |
- |
- |
PREDICTED: protein PHYTOCHROME KINASE SUBSTRATE 3 [Jatropha curcas] |
| 14 |
Hb_004316_020 |
0.1605096674 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1-like [Jatropha curcas] |
| 15 |
Hb_000656_320 |
0.164027606 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 3-like [Jatropha curcas] |
| 16 |
Hb_001541_190 |
0.1641286929 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 17 |
Hb_002200_020 |
0.164469141 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein RCOM_0137680 [Ricinus communis] |
| 18 |
Hb_003502_040 |
0.1693252496 |
- |
- |
PREDICTED: pathogen-related protein-like [Jatropha curcas] |
| 19 |
Hb_047659_010 |
0.1707119412 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004005_060 |
0.1710965855 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |