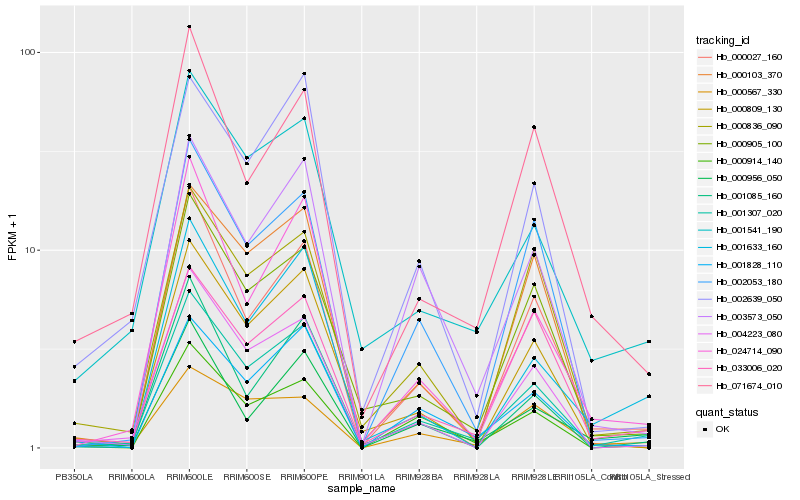
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001307_020 |
0.0 |
- |
- |
electron carrier, putative [Ricinus communis] |
| 2 |
Hb_004223_080 |
0.1083460145 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_001541_190 |
0.1096909856 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 4 |
Hb_000027_160 |
0.112589345 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 5 |
Hb_001828_110 |
0.1170830511 |
- |
- |
PREDICTED: lysine histidine transporter 1-like [Tarenaya hassleriana] |
| 6 |
Hb_000905_100 |
0.1175405388 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000809_130 |
0.1215466119 |
- |
- |
PREDICTED: peroxidase 10 isoform X3 [Jatropha curcas] |
| 8 |
Hb_000914_140 |
0.1271115502 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_024714_090 |
0.1272728371 |
rubber biosynthesis |
Gene Name: SRPP3 |
PREDICTED: stress-related protein [Jatropha curcas] |
| 10 |
Hb_003573_050 |
0.1306904199 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 11 |
Hb_002639_050 |
0.1314113715 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 12 |
Hb_001633_160 |
0.132439395 |
- |
- |
PREDICTED: zerumbone synthase-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_033006_020 |
0.132969714 |
- |
- |
Aspartate aminotransferase, putative [Ricinus communis] |
| 14 |
Hb_000836_090 |
0.1330140047 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 15 |
Hb_002053_180 |
0.1340360672 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 16 |
Hb_001085_160 |
0.1364059872 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 17 |
Hb_000103_370 |
0.1416107651 |
- |
- |
PREDICTED: protein argonaute 10 [Jatropha curcas] |
| 18 |
Hb_071674_010 |
0.141663313 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 19 |
Hb_000567_330 |
0.1426115982 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |
| 20 |
Hb_000956_050 |
0.1428102818 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |