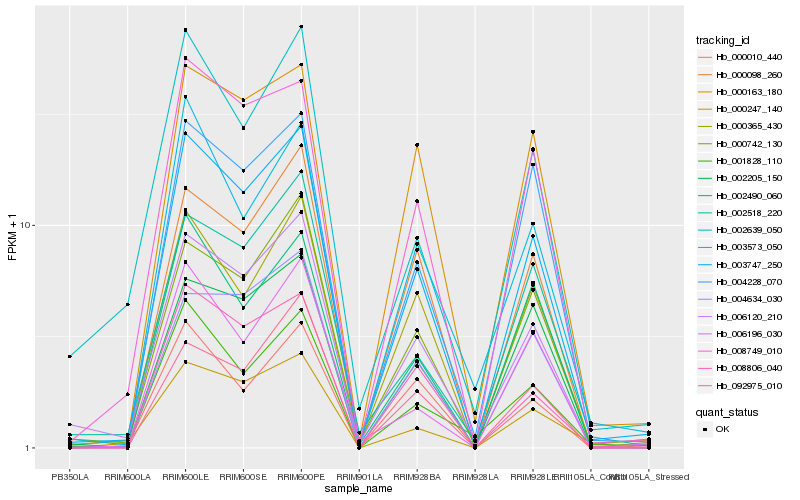
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003747_250 |
0.0 |
- |
- |
PREDICTED: potassium transporter 7 isoform X2 [Elaeis guineensis] |
| 2 |
Hb_006120_210 |
0.0814445944 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 3 |
Hb_008749_010 |
0.087991307 |
- |
- |
PREDICTED: auxin efflux carrier component 3-like [Jatropha curcas] |
| 4 |
Hb_000742_130 |
0.0962304474 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g30570 [Jatropha curcas] |
| 5 |
Hb_008806_040 |
0.0988882546 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 6 |
Hb_004228_070 |
0.1016820815 |
- |
- |
PREDICTED: uncharacterized protein LOC103338699 [Prunus mume] |
| 7 |
Hb_000365_430 |
0.1051315928 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g08570-like [Jatropha curcas] |
| 8 |
Hb_001828_110 |
0.106893736 |
- |
- |
PREDICTED: lysine histidine transporter 1-like [Tarenaya hassleriana] |
| 9 |
Hb_000163_180 |
0.1071972648 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 10 |
Hb_000098_260 |
0.1083609007 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 11 |
Hb_002490_060 |
0.1121112473 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 12 |
Hb_002518_220 |
0.1140522189 |
- |
- |
PREDICTED: uncharacterized protein LOC105631001 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004634_030 |
0.1164494356 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003573_050 |
0.1170808559 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 15 |
Hb_002205_150 |
0.1175532263 |
- |
- |
RING-H2 finger protein ATL3C, putative [Ricinus communis] |
| 16 |
Hb_006196_030 |
0.1200750318 |
- |
- |
PREDICTED: uncharacterized protein LOC105650679 [Jatropha curcas] |
| 17 |
Hb_000247_140 |
0.1204391813 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT3 [Populus euphratica] |
| 18 |
Hb_002639_050 |
0.1241353696 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 19 |
Hb_092975_010 |
0.1252379207 |
- |
- |
hypothetical protein JCGZ_04359 [Jatropha curcas] |
| 20 |
Hb_000010_440 |
0.1270328372 |
- |
- |
PREDICTED: peroxisome biogenesis factor 10-like [Jatropha curcas] |