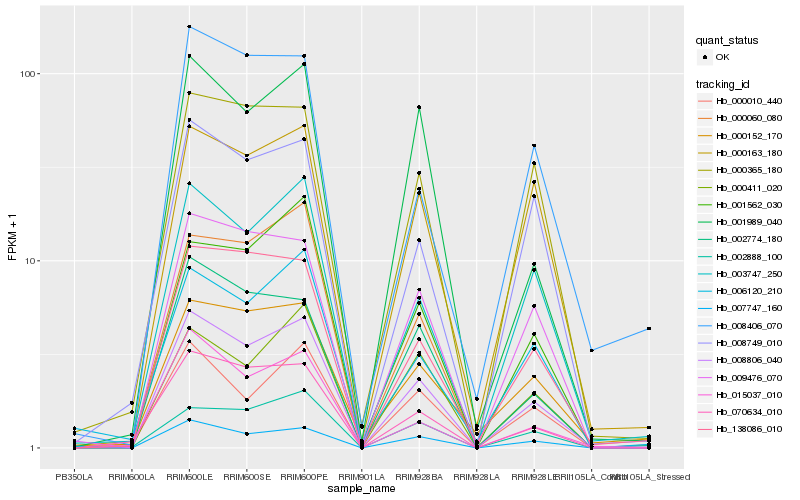
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008806_040 |
0.0 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 2 |
Hb_003747_250 |
0.0988882546 |
- |
- |
PREDICTED: potassium transporter 7 isoform X2 [Elaeis guineensis] |
| 3 |
Hb_138086_010 |
0.1069939727 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 4 |
Hb_070634_010 |
0.1098163106 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 5 |
Hb_009476_070 |
0.1099262695 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 6 |
Hb_002774_180 |
0.1157038931 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 7 |
Hb_001989_040 |
0.1208918648 |
- |
- |
PREDICTED: neurofilament heavy polypeptide [Jatropha curcas] |
| 8 |
Hb_008749_010 |
0.1233639276 |
- |
- |
PREDICTED: auxin efflux carrier component 3-like [Jatropha curcas] |
| 9 |
Hb_001562_030 |
0.1235741986 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH35 isoform X1 [Jatropha curcas] |
| 10 |
Hb_015037_010 |
0.1239710966 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 11 |
Hb_002888_100 |
0.1245598407 |
- |
- |
PREDICTED: uncharacterized protein LOC103699074, partial [Phoenix dactylifera] |
| 12 |
Hb_007747_160 |
0.1256320277 |
- |
- |
lysosomal pro-X carboxypeptidase, putative [Ricinus communis] |
| 13 |
Hb_000163_180 |
0.1269997894 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 14 |
Hb_000365_180 |
0.1271923831 |
- |
- |
PREDICTED: protein LURP-one-related 8-like [Pyrus x bretschneideri] |
| 15 |
Hb_000010_440 |
0.1291052318 |
- |
- |
PREDICTED: peroxisome biogenesis factor 10-like [Jatropha curcas] |
| 16 |
Hb_000152_170 |
0.1318690506 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 17 |
Hb_006120_210 |
0.1325784549 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 18 |
Hb_008406_070 |
0.1342002524 |
- |
- |
PREDICTED: probable protein phosphatase 2C 58 [Jatropha curcas] |
| 19 |
Hb_000411_020 |
0.1349279543 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_000060_080 |
0.137943965 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 2 protein, putative [Ricinus communis] |