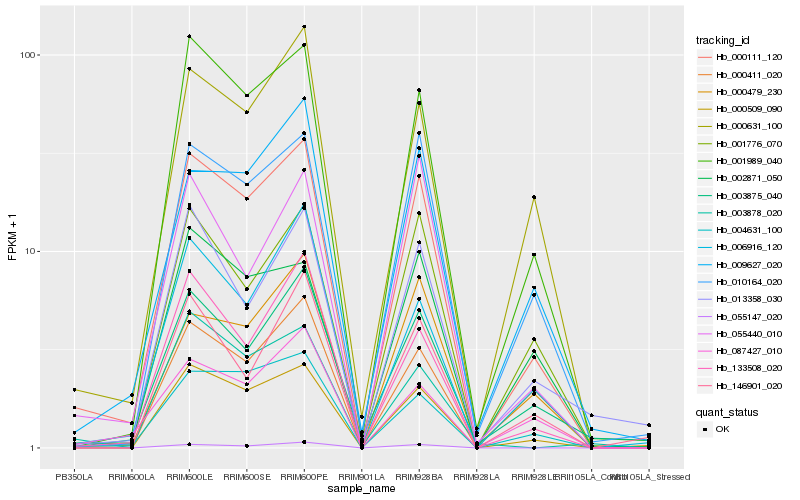
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000111_120 |
0.0 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 2 |
Hb_000509_090 |
0.0768869415 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 3 |
Hb_001989_040 |
0.0885190891 |
- |
- |
PREDICTED: neurofilament heavy polypeptide [Jatropha curcas] |
| 4 |
Hb_004631_100 |
0.1110891685 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH96 [Jatropha curcas] |
| 5 |
Hb_010164_020 |
0.114961381 |
- |
- |
PREDICTED: abscisate beta-glucosyltransferase-like [Jatropha curcas] |
| 6 |
Hb_000631_100 |
0.1153728197 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3-like [Jatropha curcas] |
| 7 |
Hb_001776_070 |
0.1240935774 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase isoform X2 [Jatropha curcas] |
| 8 |
Hb_133508_020 |
0.1254113355 |
- |
- |
PREDICTED: L-ascorbate oxidase homolog [Jatropha curcas] |
| 9 |
Hb_003875_040 |
0.1290213371 |
- |
- |
PREDICTED: endoglucanase 11-like [Jatropha curcas] |
| 10 |
Hb_055147_020 |
0.1327586026 |
- |
- |
hypothetical protein EUGRSUZ_C03837 [Eucalyptus grandis] |
| 11 |
Hb_009627_020 |
0.1361787806 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 34 [Jatropha curcas] |
| 12 |
Hb_006916_120 |
0.1369962551 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 1 [Jatropha curcas] |
| 13 |
Hb_013358_030 |
0.1426927638 |
- |
- |
PREDICTED: U-box domain-containing protein 12-like [Jatropha curcas] |
| 14 |
Hb_087427_010 |
0.1462440023 |
- |
- |
hypothetical protein PRUPE_ppa007748mg [Prunus persica] |
| 15 |
Hb_000479_230 |
0.1462635604 |
- |
- |
hypothetical protein JCGZ_19903 [Jatropha curcas] |
| 16 |
Hb_002871_050 |
0.1468146937 |
transcription factor |
TF Family: ARF |
auxin response factor 1 family protein [Populus trichocarpa] |
| 17 |
Hb_000411_020 |
0.1477985664 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 18 |
Hb_146901_020 |
0.1542094286 |
- |
- |
- |
| 19 |
Hb_003878_020 |
0.1558651065 |
- |
- |
Epidermis-specific secreted glycoprotein EP1 precursor, putative [Ricinus communis] |
| 20 |
Hb_055440_010 |
0.1574195425 |
- |
- |
protein binding protein, putative [Ricinus communis] |