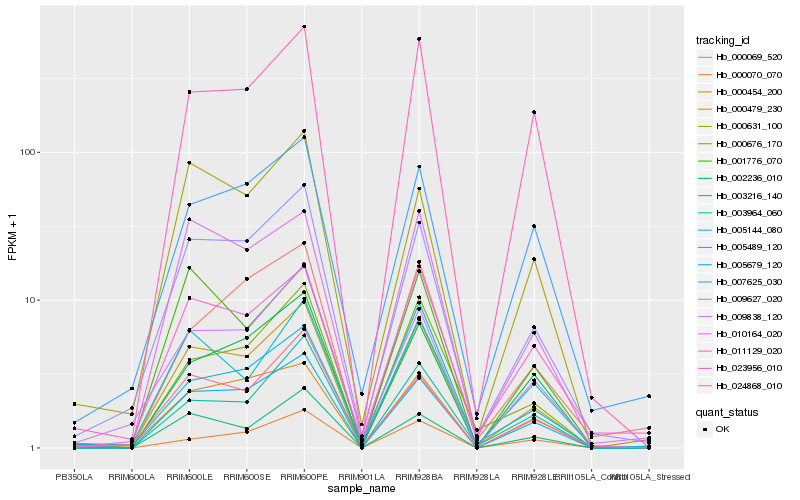
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000479_230 |
0.0 |
- |
- |
hypothetical protein JCGZ_19903 [Jatropha curcas] |
| 2 |
Hb_009627_020 |
0.0788404156 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 34 [Jatropha curcas] |
| 3 |
Hb_005679_120 |
0.0820438368 |
- |
- |
hypothetical protein RCOM_0284080 [Ricinus communis] |
| 4 |
Hb_010164_020 |
0.1111440955 |
- |
- |
PREDICTED: abscisate beta-glucosyltransferase-like [Jatropha curcas] |
| 5 |
Hb_023956_010 |
0.1120251451 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 6 |
Hb_000676_170 |
0.11233836 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2 [Jatropha curcas] |
| 7 |
Hb_003216_140 |
0.1133720829 |
- |
- |
F-box protein SNE, putative [Ricinus communis] |
| 8 |
Hb_011129_020 |
0.1139282206 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 9 |
Hb_009838_120 |
0.1176264265 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA13 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005144_080 |
0.1230360804 |
- |
- |
Serine carboxypeptidase, putative [Ricinus communis] |
| 11 |
Hb_003964_060 |
0.1242200973 |
- |
- |
PREDICTED: beta-glucosidase 40 [Jatropha curcas] |
| 12 |
Hb_005489_120 |
0.1245505003 |
- |
- |
leucine rich repeat receptor kinase, putative [Ricinus communis] |
| 13 |
Hb_000454_200 |
0.1248805481 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG7 [Jatropha curcas] |
| 14 |
Hb_024868_010 |
0.128025967 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 15 |
Hb_002236_010 |
0.1303338665 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET4 [Jatropha curcas] |
| 16 |
Hb_007625_030 |
0.1310545301 |
- |
- |
unknown [Medicago truncatula] |
| 17 |
Hb_000069_520 |
0.1373243078 |
- |
- |
PREDICTED: transcription factor TRY-like [Jatropha curcas] |
| 18 |
Hb_000631_100 |
0.1383863537 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3-like [Jatropha curcas] |
| 19 |
Hb_000070_070 |
0.1389782532 |
- |
- |
PREDICTED: protein LURP-one-related 17 [Jatropha curcas] |
| 20 |
Hb_001776_070 |
0.1397343678 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase isoform X2 [Jatropha curcas] |