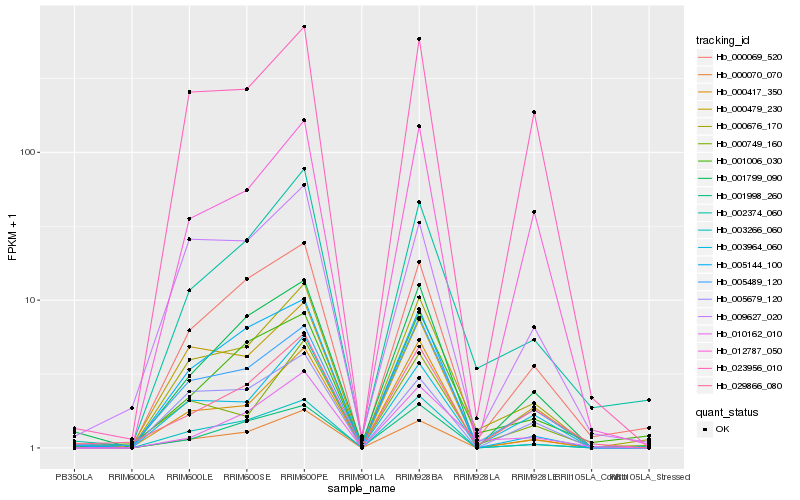
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000676_170 |
0.0 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2 [Jatropha curcas] |
| 2 |
Hb_002374_060 |
0.0967116918 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 3 |
Hb_005489_120 |
0.1068211654 |
- |
- |
leucine rich repeat receptor kinase, putative [Ricinus communis] |
| 4 |
Hb_000479_230 |
0.11233836 |
- |
- |
hypothetical protein JCGZ_19903 [Jatropha curcas] |
| 5 |
Hb_000069_520 |
0.1141767441 |
- |
- |
PREDICTED: transcription factor TRY-like [Jatropha curcas] |
| 6 |
Hb_003964_060 |
0.1150028803 |
- |
- |
PREDICTED: beta-glucosidase 40 [Jatropha curcas] |
| 7 |
Hb_003266_060 |
0.116978339 |
- |
- |
PREDICTED: basic 7S globulin-like [Populus euphratica] |
| 8 |
Hb_000749_160 |
0.1245528518 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 9 |
Hb_000070_070 |
0.1257071155 |
- |
- |
PREDICTED: protein LURP-one-related 17 [Jatropha curcas] |
| 10 |
Hb_001006_030 |
0.1260204746 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_012787_050 |
0.1365126331 |
- |
- |
PREDICTED: glutathione S-transferase U18-like [Jatropha curcas] |
| 12 |
Hb_001799_090 |
0.1424642619 |
- |
- |
PREDICTED: uclacyanin-2-like [Jatropha curcas] |
| 13 |
Hb_023956_010 |
0.1435015013 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 14 |
Hb_010162_010 |
0.1438618409 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 15 |
Hb_029866_080 |
0.144531217 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 16 |
Hb_000417_350 |
0.1472377382 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_009627_020 |
0.1507040216 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 34 [Jatropha curcas] |
| 18 |
Hb_001998_260 |
0.1518226809 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_005144_100 |
0.1526366878 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 20 |
Hb_005679_120 |
0.1532281838 |
- |
- |
hypothetical protein RCOM_0284080 [Ricinus communis] |