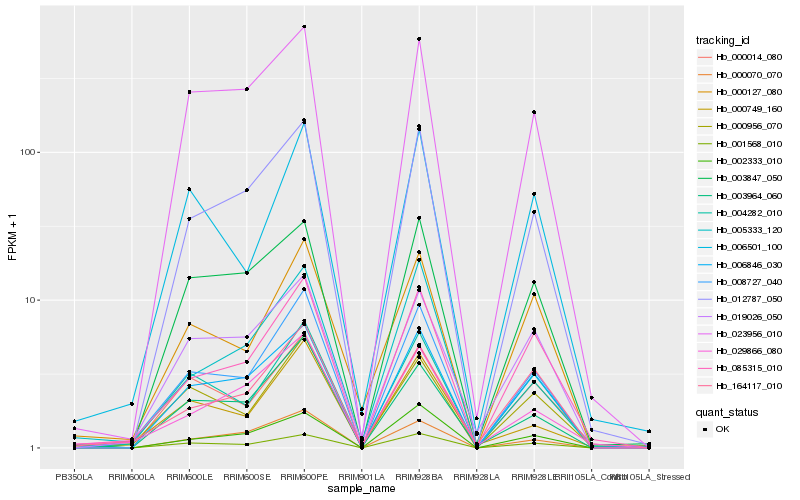
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008727_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004282_010 |
0.0874526992 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 3 |
Hb_012787_050 |
0.0905573861 |
- |
- |
PREDICTED: glutathione S-transferase U18-like [Jatropha curcas] |
| 4 |
Hb_001568_010 |
0.0909403767 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_085315_010 |
0.1022185687 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 6 |
Hb_000956_070 |
0.106964278 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 7 |
Hb_006846_030 |
0.1082096556 |
- |
- |
respiratory burst oxidase A [Manihot esculenta] |
| 8 |
Hb_003964_060 |
0.1118118547 |
- |
- |
PREDICTED: beta-glucosidase 40 [Jatropha curcas] |
| 9 |
Hb_000070_070 |
0.1128799067 |
- |
- |
PREDICTED: protein LURP-one-related 17 [Jatropha curcas] |
| 10 |
Hb_023956_010 |
0.1148323403 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 11 |
Hb_006501_100 |
0.120838869 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 12 |
Hb_000014_080 |
0.1211913324 |
- |
- |
PREDICTED: UDP-glycosyltransferase 87A1-like [Jatropha curcas] |
| 13 |
Hb_002333_010 |
0.1255792412 |
- |
- |
hypothetical protein JCGZ_18834 [Jatropha curcas] |
| 14 |
Hb_000127_080 |
0.1264970173 |
- |
- |
hypothetical protein POPTR_0005s21760g [Populus trichocarpa] |
| 15 |
Hb_019026_050 |
0.1266250231 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000749_160 |
0.1269085873 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 17 |
Hb_164117_010 |
0.1280675471 |
- |
- |
hypothetical protein PHAVU_004G0480000g [Phaseolus vulgaris] |
| 18 |
Hb_005333_120 |
0.1341456934 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Jatropha curcas] |
| 19 |
Hb_029866_080 |
0.1346778325 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 20 |
Hb_003847_050 |
0.1348733202 |
- |
- |
PREDICTED: aquaporin SIP1-1 [Jatropha curcas] |