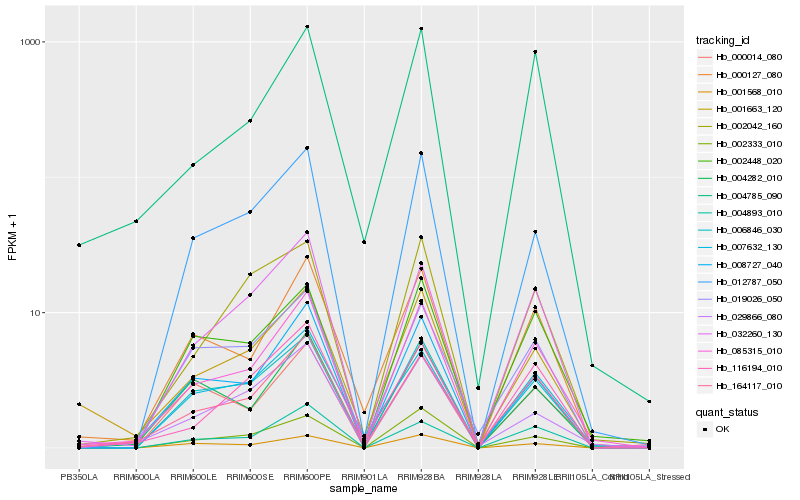
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_085315_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 2 |
Hb_164117_010 |
0.089729491 |
- |
- |
hypothetical protein PHAVU_004G0480000g [Phaseolus vulgaris] |
| 3 |
Hb_008727_040 |
0.1022185687 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_006846_030 |
0.1080252604 |
- |
- |
respiratory burst oxidase A [Manihot esculenta] |
| 5 |
Hb_012787_050 |
0.1159032291 |
- |
- |
PREDICTED: glutathione S-transferase U18-like [Jatropha curcas] |
| 6 |
Hb_004893_010 |
0.1165034675 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Jatropha curcas] |
| 7 |
Hb_000127_080 |
0.1177971693 |
- |
- |
hypothetical protein POPTR_0005s21760g [Populus trichocarpa] |
| 8 |
Hb_032260_130 |
0.1179300154 |
- |
- |
hypothetical protein POPTR_0003s13800g [Populus trichocarpa] |
| 9 |
Hb_001568_010 |
0.125481515 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002448_020 |
0.1301236872 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 11 |
Hb_002333_010 |
0.1306805019 |
- |
- |
hypothetical protein JCGZ_18834 [Jatropha curcas] |
| 12 |
Hb_004282_010 |
0.1309773096 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 13 |
Hb_001663_120 |
0.1327351949 |
- |
- |
PREDICTED: formin-like protein 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007632_130 |
0.1333406819 |
- |
- |
PREDICTED: nudix hydrolase 10-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_004785_090 |
0.135867169 |
- |
- |
hypothetical protein MIMGU_mgv1a018797mg [Erythranthe guttata] |
| 16 |
Hb_002042_160 |
0.1368522917 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 17 |
Hb_116194_010 |
0.1387886755 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 18 |
Hb_029866_080 |
0.1387893496 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 19 |
Hb_019026_050 |
0.1390855701 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000014_080 |
0.1390954057 |
- |
- |
PREDICTED: UDP-glycosyltransferase 87A1-like [Jatropha curcas] |