| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
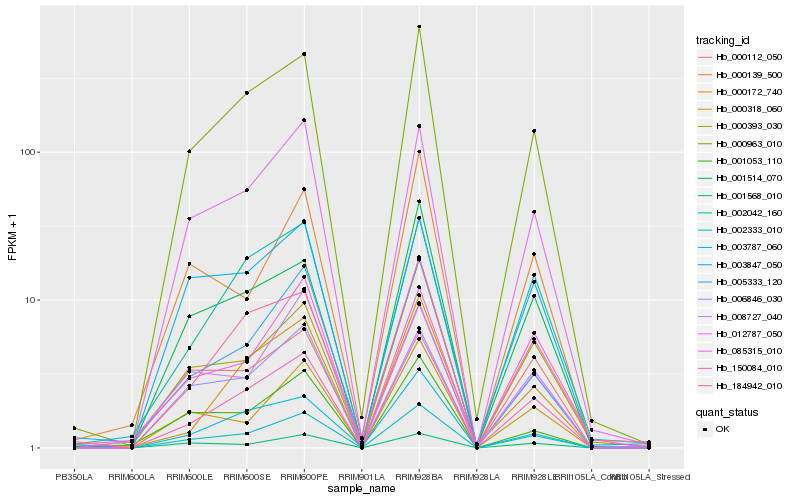
Hb_002333_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_18834 [Jatropha curcas] |
| 2 |
Hb_150084_010 |
0.0540598686 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Vitis vinifera] |
| 3 |
Hb_000963_010 |
0.065893381 |
- |
- |
PREDICTED: aquaporin PIP2-4 [Jatropha curcas] |
| 4 |
Hb_012787_050 |
0.0802449407 |
- |
- |
PREDICTED: glutathione S-transferase U18-like [Jatropha curcas] |
| 5 |
Hb_001568_010 |
0.0993076297 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000318_060 |
0.0996680484 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005333_120 |
0.1071435017 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Jatropha curcas] |
| 8 |
Hb_006846_030 |
0.1074124597 |
- |
- |
respiratory burst oxidase A [Manihot esculenta] |
| 9 |
Hb_000393_030 |
0.108624937 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 10 |
Hb_003847_050 |
0.1225177248 |
- |
- |
PREDICTED: aquaporin SIP1-1 [Jatropha curcas] |
| 11 |
Hb_001514_070 |
0.1227156306 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_001053_110 |
0.123881586 |
- |
- |
PREDICTED: uncharacterized protein LOC105628699 [Jatropha curcas] |
| 13 |
Hb_003787_060 |
0.1240535725 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 14 |
Hb_184942_010 |
0.1248101351 |
- |
- |
STS14 protein precursor, putative [Ricinus communis] |
| 15 |
Hb_000112_050 |
0.1255566457 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1 [Jatropha curcas] |
| 16 |
Hb_008727_040 |
0.1255792412 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000139_500 |
0.1271160115 |
- |
- |
PREDICTED: putative uncharacterized protein DDB_G0286901 [Prunus mume] |
| 18 |
Hb_002042_160 |
0.1285302986 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 19 |
Hb_000172_740 |
0.1304923174 |
- |
- |
HXXXD-type acyl-transferase family protein, putative [Theobroma cacao] |
| 20 |
Hb_085315_010 |
0.1306805019 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |