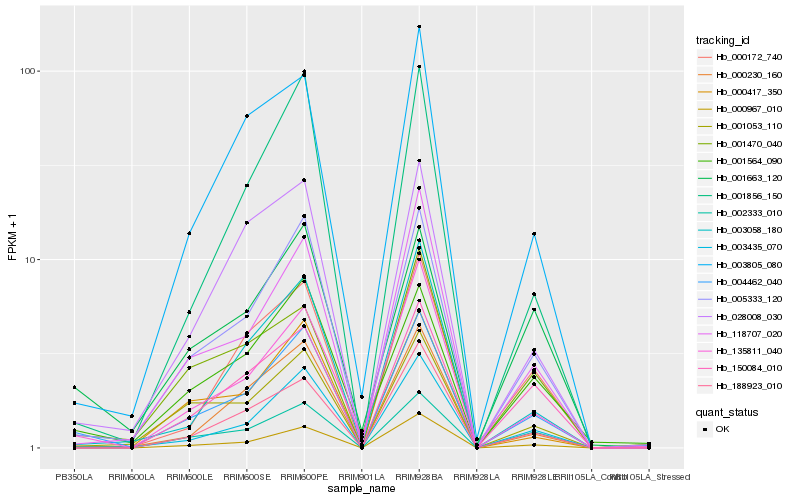
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004462_040 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_005333_120 |
0.0808482501 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Jatropha curcas] |
| 3 |
Hb_003435_070 |
0.1056586525 |
- |
- |
PREDICTED: probable pectate lyase 12 isoform X3 [Jatropha curcas] |
| 4 |
Hb_135811_040 |
0.1258988871 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001053_110 |
0.1261322359 |
- |
- |
PREDICTED: uncharacterized protein LOC105628699 [Jatropha curcas] |
| 6 |
Hb_118707_020 |
0.1284847231 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001564_090 |
0.1318203966 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002333_010 |
0.1321481341 |
- |
- |
hypothetical protein JCGZ_18834 [Jatropha curcas] |
| 9 |
Hb_001856_150 |
0.1324906465 |
- |
- |
- |
| 10 |
Hb_000967_010 |
0.1340699011 |
- |
- |
hypothetical protein JCGZ_08336 [Jatropha curcas] |
| 11 |
Hb_028008_030 |
0.1352907955 |
- |
- |
probable xyloglucan endotransglucosylase/hydrolase protein 23 precursor [Malus domestica] |
| 12 |
Hb_001663_120 |
0.1357258681 |
- |
- |
PREDICTED: formin-like protein 1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000230_160 |
0.1383672289 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003805_080 |
0.139582336 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 15 |
Hb_000417_350 |
0.1413907678 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_188923_010 |
0.143371593 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 17 |
Hb_150084_010 |
0.1437424846 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Vitis vinifera] |
| 18 |
Hb_003058_180 |
0.1450881873 |
transcription factor |
TF Family: HSF |
hypothetical protein POPTR_0001s28040g [Populus trichocarpa] |
| 19 |
Hb_000172_740 |
0.1462327669 |
- |
- |
HXXXD-type acyl-transferase family protein, putative [Theobroma cacao] |
| 20 |
Hb_001470_040 |
0.1464296152 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 3 [Jatropha curcas] |