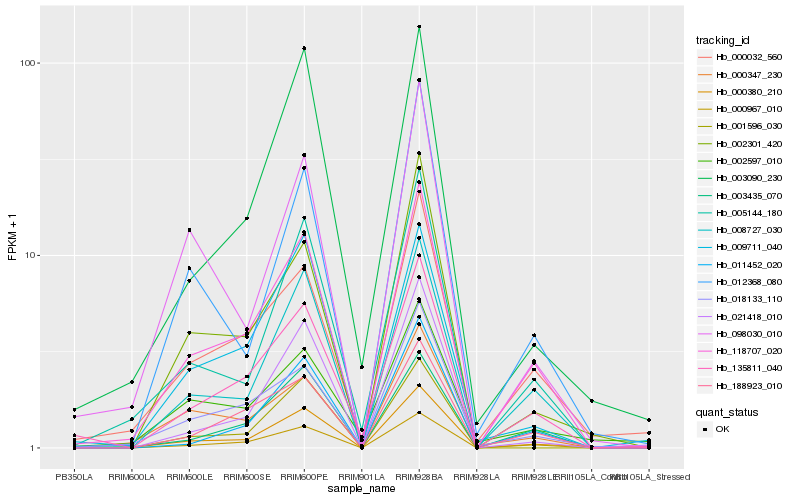
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000380_210 |
0.0 |
- |
- |
Cyclic nucleotide-gated channel 15 [Theobroma cacao] |
| 2 |
Hb_118707_020 |
0.1069590791 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000967_010 |
0.1133923082 |
- |
- |
hypothetical protein JCGZ_08336 [Jatropha curcas] |
| 4 |
Hb_021418_010 |
0.1189463812 |
- |
- |
PREDICTED: probable pectate lyase 12 [Prunus mume] |
| 5 |
Hb_008727_030 |
0.1194257633 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_018133_110 |
0.1221936832 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_000032_560 |
0.1224001687 |
- |
- |
hypothetical protein JCGZ_15037 [Jatropha curcas] |
| 8 |
Hb_005144_180 |
0.1238617376 |
- |
- |
PREDICTED: protein enabled homolog [Populus euphratica] |
| 9 |
Hb_003090_230 |
0.1295709172 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002301_420 |
0.1322077499 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0011s05320g [Populus trichocarpa] |
| 11 |
Hb_135811_040 |
0.1408231975 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_009711_040 |
0.141161245 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |
| 13 |
Hb_000347_230 |
0.1430073306 |
- |
- |
inter-alpha-trypsin inhibitor heavy chain, putative [Ricinus communis] |
| 14 |
Hb_011452_020 |
0.144182684 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 4 [Jatropha curcas] |
| 15 |
Hb_002597_010 |
0.1447404515 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Malus domestica] |
| 16 |
Hb_003435_070 |
0.1473522759 |
- |
- |
PREDICTED: probable pectate lyase 12 isoform X3 [Jatropha curcas] |
| 17 |
Hb_012368_080 |
0.1496293774 |
- |
- |
PREDICTED: uncharacterized protein LOC105126277 [Populus euphratica] |
| 18 |
Hb_188923_010 |
0.1501186202 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 19 |
Hb_001596_030 |
0.1514312696 |
- |
- |
PREDICTED: uncharacterized protein LOC105141672 [Populus euphratica] |
| 20 |
Hb_098030_010 |
0.1520528488 |
- |
- |
hypothetical protein JCGZ_06251 [Jatropha curcas] |