| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
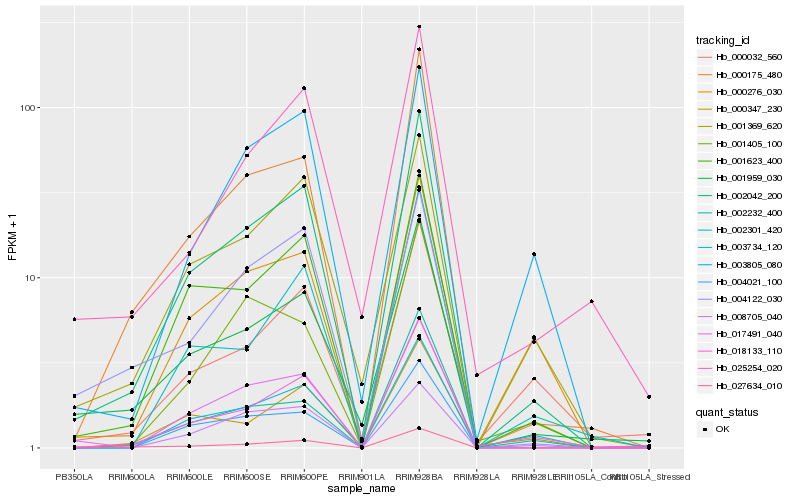
Hb_002042_200 |
0.0 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_001623_400 |
0.0791566136 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_018133_110 |
0.0965818234 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_000175_480 |
0.0967114002 |
- |
- |
hypothetical protein JCGZ_06165 [Jatropha curcas] |
| 5 |
Hb_027634_010 |
0.0983454877 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_017491_040 |
0.1063830123 |
- |
- |
PREDICTED: uncharacterized protein LOC105631348 [Jatropha curcas] |
| 7 |
Hb_001959_030 |
0.1094780755 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003734_120 |
0.1118906542 |
- |
- |
PREDICTED: uncharacterized protein LOC105632812 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004021_100 |
0.116781972 |
- |
- |
- |
| 10 |
Hb_001369_620 |
0.1273463802 |
- |
- |
PREDICTED: protein EXORDIUM-like 3 [Jatropha curcas] |
| 11 |
Hb_002301_420 |
0.1308266479 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0011s05320g [Populus trichocarpa] |
| 12 |
Hb_000276_030 |
0.1323780328 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 13 |
Hb_000347_230 |
0.1348850451 |
- |
- |
inter-alpha-trypsin inhibitor heavy chain, putative [Ricinus communis] |
| 14 |
Hb_002232_400 |
0.1394815185 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH25-like [Jatropha curcas] |
| 15 |
Hb_008705_040 |
0.1426895695 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 16 |
Hb_025254_020 |
0.1429314448 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Vitis vinifera] |
| 17 |
Hb_004122_030 |
0.1434276992 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 4-like [Populus euphratica] |
| 18 |
Hb_000032_560 |
0.1462216418 |
- |
- |
hypothetical protein JCGZ_15037 [Jatropha curcas] |
| 19 |
Hb_001405_100 |
0.1479353581 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 4 [Jatropha curcas] |
| 20 |
Hb_003805_080 |
0.1490795801 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |