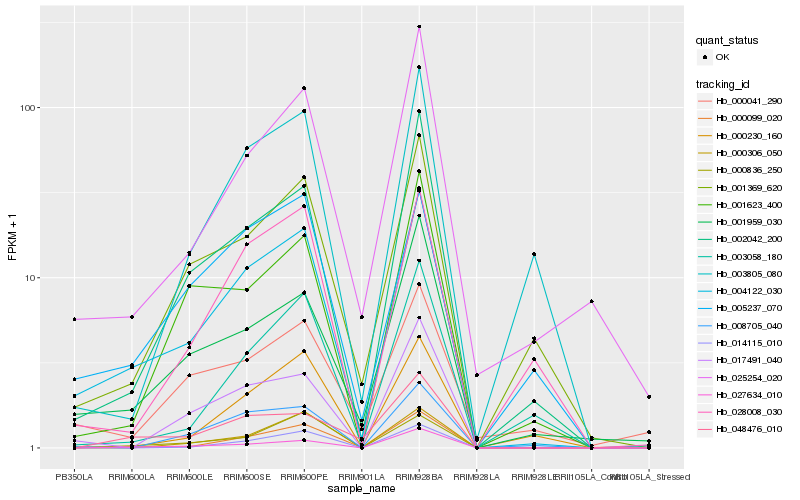
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004122_030 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 4-like [Populus euphratica] |
| 2 |
Hb_002042_200 |
0.1434276992 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_000099_020 |
0.1437196994 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: uncharacterized protein LOC103431057 [Malus domestica] |
| 4 |
Hb_027634_010 |
0.1462419662 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_001959_030 |
0.1487195202 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001369_620 |
0.1505956145 |
- |
- |
PREDICTED: protein EXORDIUM-like 3 [Jatropha curcas] |
| 7 |
Hb_005237_070 |
0.1549537867 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 4-like [Populus euphratica] |
| 8 |
Hb_017491_040 |
0.1574357354 |
- |
- |
PREDICTED: uncharacterized protein LOC105631348 [Jatropha curcas] |
| 9 |
Hb_008705_040 |
0.1595802623 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 10 |
Hb_025254_020 |
0.1660303421 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Vitis vinifera] |
| 11 |
Hb_028008_030 |
0.168417681 |
- |
- |
probable xyloglucan endotransglucosylase/hydrolase protein 23 precursor [Malus domestica] |
| 12 |
Hb_000836_250 |
0.1691971748 |
- |
- |
hypothetical protein VITISV_022299 [Vitis vinifera] |
| 13 |
Hb_001623_400 |
0.1747778123 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003805_080 |
0.1785437718 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 15 |
Hb_048476_010 |
0.1790153684 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Citrus sinensis] |
| 16 |
Hb_000041_290 |
0.1823881217 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 17 |
Hb_000230_160 |
0.1832387378 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003058_180 |
0.1838047852 |
transcription factor |
TF Family: HSF |
hypothetical protein POPTR_0001s28040g [Populus trichocarpa] |
| 19 |
Hb_014115_010 |
0.1846609889 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 20 |
Hb_000306_050 |
0.1856826201 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |