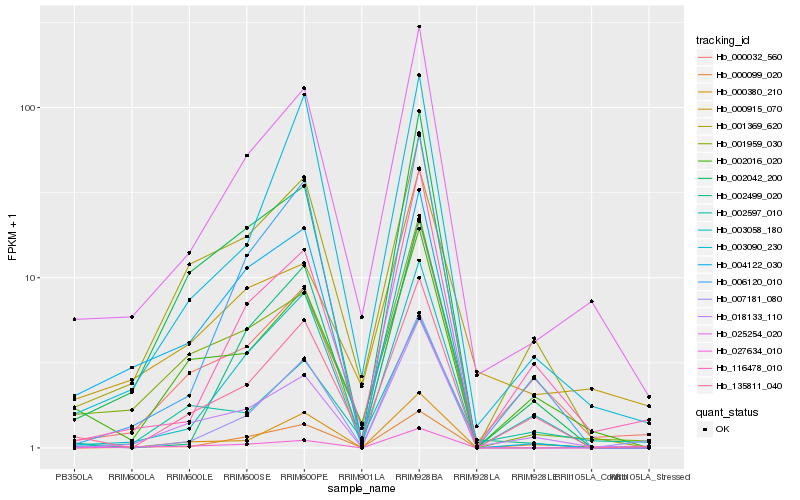
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025254_020 |
0.0 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Vitis vinifera] |
| 2 |
Hb_000099_020 |
0.1169717461 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: uncharacterized protein LOC103431057 [Malus domestica] |
| 3 |
Hb_001959_030 |
0.1234362402 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_018133_110 |
0.129497893 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_002042_200 |
0.1429314448 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_000032_560 |
0.1431531476 |
- |
- |
hypothetical protein JCGZ_15037 [Jatropha curcas] |
| 7 |
Hb_002016_020 |
0.1460574898 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 8 |
Hb_135811_040 |
0.1469348441 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_116478_010 |
0.1489214615 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF003-like [Jatropha curcas] |
| 10 |
Hb_000915_070 |
0.1528783578 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_006120_010 |
0.1547456916 |
- |
- |
invertase inhibitor [Manihot esculenta] |
| 12 |
Hb_003058_180 |
0.1547749678 |
transcription factor |
TF Family: HSF |
hypothetical protein POPTR_0001s28040g [Populus trichocarpa] |
| 13 |
Hb_002597_010 |
0.1569548681 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Malus domestica] |
| 14 |
Hb_001369_620 |
0.1586895986 |
- |
- |
PREDICTED: protein EXORDIUM-like 3 [Jatropha curcas] |
| 15 |
Hb_003090_230 |
0.1600706558 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000380_210 |
0.164012187 |
- |
- |
Cyclic nucleotide-gated channel 15 [Theobroma cacao] |
| 17 |
Hb_002499_020 |
0.1655368736 |
- |
- |
hevamine-A precursor, putative [Ricinus communis] |
| 18 |
Hb_027634_010 |
0.1659632629 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_004122_030 |
0.1660303421 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 4-like [Populus euphratica] |
| 20 |
Hb_007181_080 |
0.1663002385 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |