| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
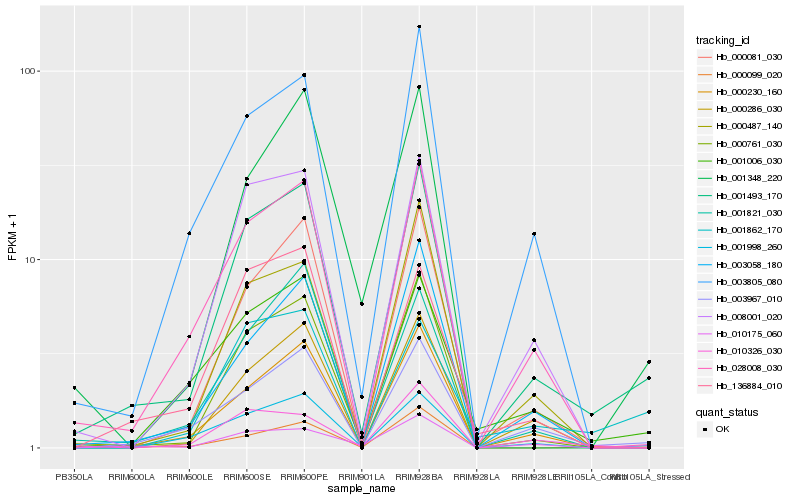
Hb_001493_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631328 [Jatropha curcas] |
| 2 |
Hb_003967_010 |
0.1288362424 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_028008_030 |
0.1395391493 |
- |
- |
probable xyloglucan endotransglucosylase/hydrolase protein 23 precursor [Malus domestica] |
| 4 |
Hb_000761_030 |
0.14065841 |
transcription factor |
TF Family: ERF |
Transcriptional factor TINY, putative [Ricinus communis] |
| 5 |
Hb_000230_160 |
0.1452154133 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X1 [Jatropha curcas] |
| 6 |
Hb_008001_020 |
0.1469857375 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT-like [Populus euphratica] |
| 7 |
Hb_000081_030 |
0.148472695 |
- |
- |
BnaC09g05810D [Brassica napus] |
| 8 |
Hb_000099_020 |
0.1503710884 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: uncharacterized protein LOC103431057 [Malus domestica] |
| 9 |
Hb_001006_030 |
0.1578804303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000286_030 |
0.160029584 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 11 |
Hb_003058_180 |
0.1634245064 |
transcription factor |
TF Family: HSF |
hypothetical protein POPTR_0001s28040g [Populus trichocarpa] |
| 12 |
Hb_000487_140 |
0.1645175618 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 13 |
Hb_003805_080 |
0.1665547352 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 14 |
Hb_001821_030 |
0.1686579484 |
- |
- |
hypothetical protein JCGZ_13357 [Jatropha curcas] |
| 15 |
Hb_001862_170 |
0.1704273974 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_001998_260 |
0.1756068387 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_136884_010 |
0.1794337223 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 18 |
Hb_010175_060 |
0.1832505249 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_010326_030 |
0.1836118214 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 4-like [Jatropha curcas] |
| 20 |
Hb_001348_220 |
0.1854145346 |
- |
- |
PREDICTED: defensin Ec-AMP-D2-like [Nelumbo nucifera] |