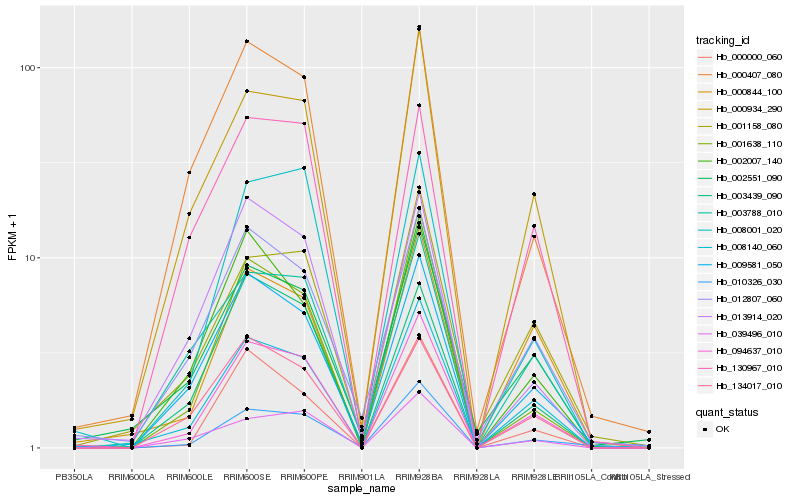
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_094637_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642042 [Jatropha curcas] |
| 2 |
Hb_008140_060 |
0.0690779429 |
- |
- |
Cytochrome P450, putative [Theobroma cacao] |
| 3 |
Hb_000934_290 |
0.0937370851 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_009581_050 |
0.1021556805 |
- |
- |
hypothetical protein CISIN_1g0025812mg, partial [Citrus sinensis] |
| 5 |
Hb_001638_110 |
0.1098376354 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000000_060 |
0.1132324811 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 14 [Jatropha curcas] |
| 7 |
Hb_039496_010 |
0.1151497642 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 8 |
Hb_008001_020 |
0.1194941647 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT-like [Populus euphratica] |
| 9 |
Hb_134017_010 |
0.1195779994 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_000844_100 |
0.1230352387 |
- |
- |
hypothetical protein JCGZ_04351 [Jatropha curcas] |
| 11 |
Hb_003439_090 |
0.1244682998 |
- |
- |
kinase, putative [Ricinus communis] |
| 12 |
Hb_002007_140 |
0.1251897807 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER-like [Populus euphratica] |
| 13 |
Hb_001158_080 |
0.1268280457 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 14 |
Hb_000407_080 |
0.1272143826 |
- |
- |
1-aminocyclopropane-1-carboxylate oxidase [Hevea brasiliensis] |
| 15 |
Hb_010326_030 |
0.1283388155 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 4-like [Jatropha curcas] |
| 16 |
Hb_003788_010 |
0.1304386381 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 17 |
Hb_012807_060 |
0.13061893 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_130967_010 |
0.13124272 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 19 |
Hb_002551_090 |
0.1312702149 |
- |
- |
hypothetical protein JCGZ_17224 [Jatropha curcas] |
| 20 |
Hb_013914_020 |
0.1319923845 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |