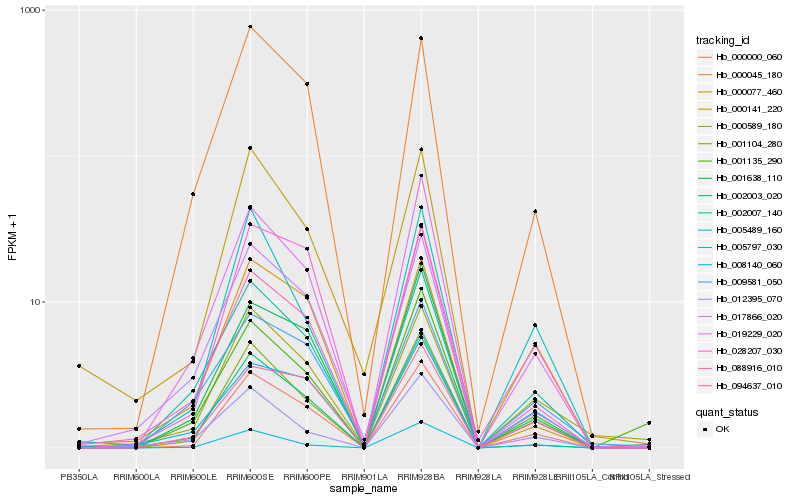
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000000_060 |
0.0 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 14 [Jatropha curcas] |
| 2 |
Hb_001104_280 |
0.0653325371 |
- |
- |
PREDICTED: UPF0481 protein At3g47200 [Jatropha curcas] |
| 3 |
Hb_019229_020 |
0.0951055276 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_001638_110 |
0.109491331 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002007_140 |
0.1122088059 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER-like [Populus euphratica] |
| 6 |
Hb_094637_010 |
0.1132324811 |
- |
- |
PREDICTED: uncharacterized protein LOC105642042 [Jatropha curcas] |
| 7 |
Hb_000045_180 |
0.1254605763 |
- |
- |
aquaporin PIP1 [Hevea brasiliensis] |
| 8 |
Hb_002003_020 |
0.1257057604 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 9 |
Hb_000589_180 |
0.1264939776 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 10 |
Hb_005489_160 |
0.1317002608 |
- |
- |
PREDICTED: primary amine oxidase-like [Jatropha curcas] |
| 11 |
Hb_017866_020 |
0.1318966272 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 40 [Jatropha curcas] |
| 12 |
Hb_000077_460 |
0.1330040366 |
- |
- |
- |
| 13 |
Hb_008140_060 |
0.1369328428 |
- |
- |
Cytochrome P450, putative [Theobroma cacao] |
| 14 |
Hb_000141_220 |
0.1373912062 |
- |
- |
PREDICTED: 14 kDa proline-rich protein DC2.15-like [Gossypium raimondii] |
| 15 |
Hb_088916_010 |
0.1393723841 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At4g25390 [Jatropha curcas] |
| 16 |
Hb_005797_030 |
0.1424258817 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like [Jatropha curcas] |
| 17 |
Hb_009581_050 |
0.1436169981 |
- |
- |
hypothetical protein CISIN_1g0025812mg, partial [Citrus sinensis] |
| 18 |
Hb_028207_030 |
0.1444139381 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 8-like [Citrus sinensis] |
| 19 |
Hb_001135_290 |
0.1472559174 |
- |
- |
hypothetical protein JCGZ_06658 [Jatropha curcas] |
| 20 |
Hb_012395_070 |
0.1493018868 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL6 [Jatropha curcas] |