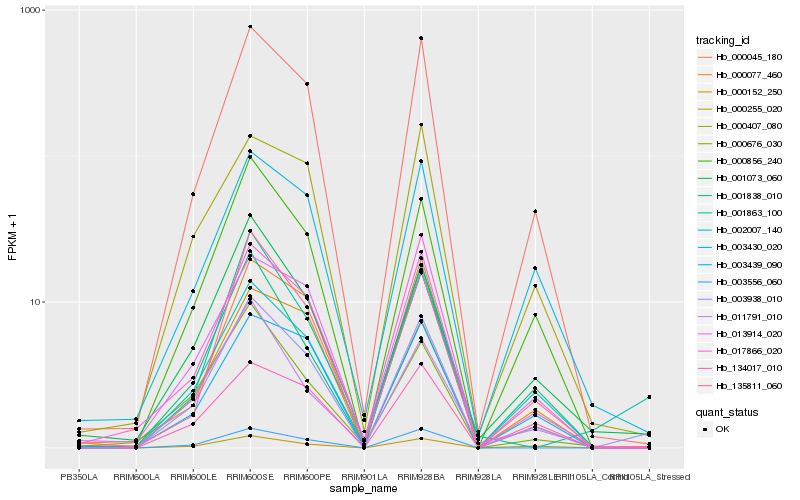
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003938_010 |
0.0 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor 1B, putative [Ricinus communis] |
| 2 |
Hb_000045_180 |
0.0947327743 |
- |
- |
aquaporin PIP1 [Hevea brasiliensis] |
| 3 |
Hb_000856_240 |
0.1101283644 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_000255_020 |
0.1262310439 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 5 |
Hb_001073_060 |
0.128273527 |
- |
- |
phosphate transporter 1 [Hevea brasiliensis] |
| 6 |
Hb_001838_010 |
0.1322526047 |
- |
- |
hypothetical protein POPTR_0005s18380g [Populus trichocarpa] |
| 7 |
Hb_135811_060 |
0.1325083405 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 8 |
Hb_003556_060 |
0.1370363747 |
- |
- |
- |
| 9 |
Hb_003439_090 |
0.1375355418 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_013914_020 |
0.1379706299 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 11 |
Hb_000077_460 |
0.1401208609 |
- |
- |
- |
| 12 |
Hb_017866_020 |
0.1411516766 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 40 [Jatropha curcas] |
| 13 |
Hb_003430_020 |
0.1444522658 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 2 member B7, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_002007_140 |
0.1458759762 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER-like [Populus euphratica] |
| 15 |
Hb_134017_010 |
0.1479679956 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_000152_250 |
0.1499743949 |
- |
- |
PREDICTED: uncharacterized protein LOC105648569 [Jatropha curcas] |
| 17 |
Hb_000407_080 |
0.1502347904 |
- |
- |
1-aminocyclopropane-1-carboxylate oxidase [Hevea brasiliensis] |
| 18 |
Hb_000676_030 |
0.151100553 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-B [Jatropha curcas] |
| 19 |
Hb_011791_010 |
0.1544461234 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 20 |
Hb_001863_100 |
0.1551654016 |
- |
- |
conserved hypothetical protein [Ricinus communis] |