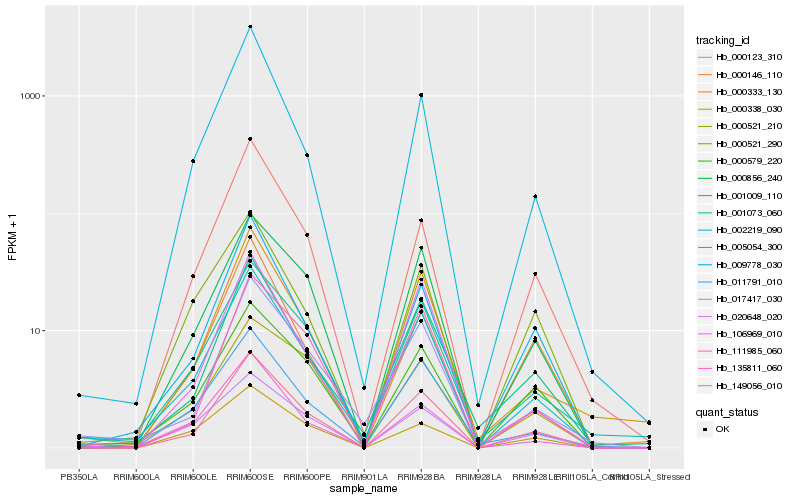
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_149056_010 |
0.0 |
- |
- |
hypothetical protein CICLE_v10016664mg [Citrus clementina] |
| 2 |
Hb_000856_240 |
0.082605513 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_002219_090 |
0.086843933 |
- |
- |
protein kinase atmrk1, putative [Ricinus communis] |
| 4 |
Hb_001009_110 |
0.1059478201 |
- |
- |
PREDICTED: uncharacterized protein LOC105640232 [Jatropha curcas] |
| 5 |
Hb_000579_220 |
0.1059529285 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_009778_030 |
0.1067891263 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP23-like [Populus euphratica] |
| 7 |
Hb_000123_310 |
0.1079418673 |
- |
- |
PREDICTED: aquaporin TIP1-3 [Jatropha curcas] |
| 8 |
Hb_000333_130 |
0.1109394975 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_005054_300 |
0.1155212594 |
- |
- |
aquaporin [Hevea brasiliensis] |
| 10 |
Hb_001073_060 |
0.1157284414 |
- |
- |
phosphate transporter 1 [Hevea brasiliensis] |
| 11 |
Hb_135811_060 |
0.1190124764 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 12 |
Hb_011791_010 |
0.1198128621 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 13 |
Hb_020648_020 |
0.1243475669 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor pho1, putative [Ricinus communis] |
| 14 |
Hb_017417_030 |
0.1259011705 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 15 |
Hb_000146_110 |
0.1321849759 |
- |
- |
resveratrol/hydroxycinnamic acid O-glucosyltransferase [Vitis labrusca] |
| 16 |
Hb_106969_010 |
0.132343934 |
- |
- |
Cytochrome P450 76C4 [Glycine soja] |
| 17 |
Hb_111985_060 |
0.1333472394 |
- |
- |
PREDICTED: cytokinin dehydrogenase 5 [Jatropha curcas] |
| 18 |
Hb_000521_210 |
0.1339329535 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 19 |
Hb_000521_290 |
0.1364860313 |
- |
- |
hypothetical protein POPTR_0001s23530g [Populus trichocarpa] |
| 20 |
Hb_000338_030 |
0.1365086593 |
transcription factor |
TF Family: HB |
hypothetical protein JCGZ_21254 [Jatropha curcas] |