| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
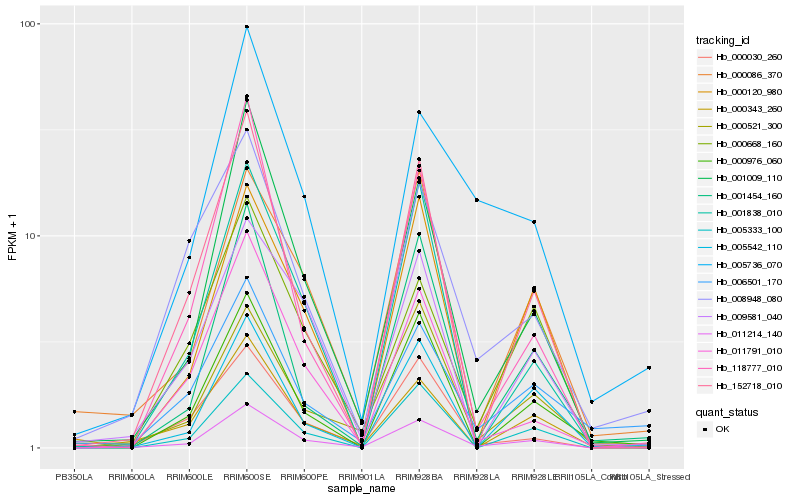
Hb_011214_140 |
0.0 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 2 |
Hb_001009_110 |
0.112535208 |
- |
- |
PREDICTED: uncharacterized protein LOC105640232 [Jatropha curcas] |
| 3 |
Hb_000030_260 |
0.142610162 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 4 |
Hb_000343_260 |
0.1477836582 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 5 |
Hb_000521_300 |
0.1479849692 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 6 |
Hb_000086_370 |
0.1483364329 |
- |
- |
Long-chain-alcohol oxidase FAO2 [Triticum urartu] |
| 7 |
Hb_006501_170 |
0.1490086262 |
- |
- |
hypothetical protein JCGZ_07371 [Jatropha curcas] |
| 8 |
Hb_152718_010 |
0.1501954652 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 9 |
Hb_005333_100 |
0.1511076442 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001454_160 |
0.1512617061 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 11 |
Hb_005542_110 |
0.1529225614 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01203 [Jatropha curcas] |
| 12 |
Hb_000976_060 |
0.1530641833 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 13 |
Hb_000668_160 |
0.1586562486 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 14 |
Hb_011791_010 |
0.1586791699 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 15 |
Hb_001838_010 |
0.1598711683 |
- |
- |
hypothetical protein POPTR_0005s18380g [Populus trichocarpa] |
| 16 |
Hb_005736_070 |
0.1604067417 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 17 |
Hb_008948_080 |
0.1615478271 |
- |
- |
PREDICTED: TBC1 domain family member 13-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_009581_040 |
0.1621177332 |
- |
- |
hypothetical protein B456_011G052100 [Gossypium raimondii] |
| 19 |
Hb_000120_980 |
0.1624659627 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 5-like [Jatropha curcas] |
| 20 |
Hb_118777_010 |
0.1644127742 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |