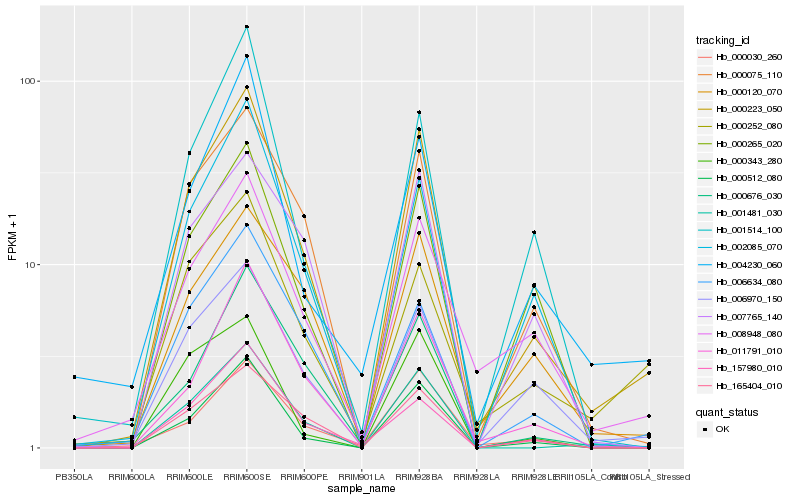
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000223_050 |
0.0 |
- |
- |
PREDICTED: endochitinase A-like [Jatropha curcas] |
| 2 |
Hb_000512_080 |
0.1058900929 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_000075_110 |
0.1203899185 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 4 |
Hb_006970_150 |
0.1216210816 |
- |
- |
PREDICTED: uncharacterized protein LOC105650471 [Jatropha curcas] |
| 5 |
Hb_001481_030 |
0.122558813 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_165404_010 |
0.1281138597 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 7 |
Hb_002085_070 |
0.1298967268 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_000030_260 |
0.13468942 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 9 |
Hb_011791_010 |
0.1375712985 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 10 |
Hb_008948_080 |
0.1406029105 |
- |
- |
PREDICTED: TBC1 domain family member 13-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000265_020 |
0.143591256 |
- |
- |
hypothetical protein POPTR_0011s16980g, partial [Populus trichocarpa] |
| 12 |
Hb_006634_080 |
0.1464022599 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b [Jatropha curcas] |
| 13 |
Hb_000120_070 |
0.1477779859 |
- |
- |
PREDICTED: uncharacterized protein LOC105630351 [Jatropha curcas] |
| 14 |
Hb_004230_060 |
0.1513622722 |
- |
- |
PREDICTED: ferritin, chloroplastic-like [Populus euphratica] |
| 15 |
Hb_000252_080 |
0.1517714639 |
- |
- |
PREDICTED: uncharacterized protein LOC105644644 [Jatropha curcas] |
| 16 |
Hb_157980_010 |
0.1524619564 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 17 |
Hb_001514_100 |
0.153827532 |
- |
- |
PREDICTED: F-box protein GID2 [Jatropha curcas] |
| 18 |
Hb_000676_030 |
0.1545009148 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-B [Jatropha curcas] |
| 19 |
Hb_000343_280 |
0.154589488 |
- |
- |
hypothetical protein JCGZ_24094 [Jatropha curcas] |
| 20 |
Hb_007765_140 |
0.1564077618 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 8.1-like [Jatropha curcas] |