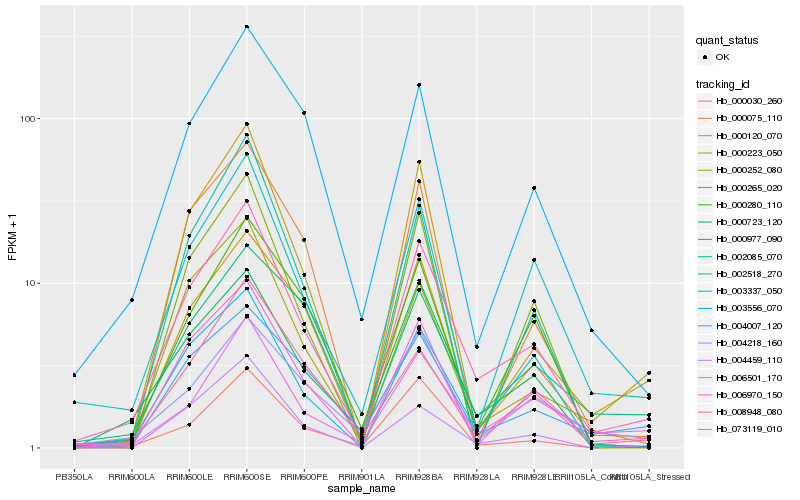
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008948_080 |
0.0 |
- |
- |
PREDICTED: TBC1 domain family member 13-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_006970_150 |
0.1120126388 |
- |
- |
PREDICTED: uncharacterized protein LOC105650471 [Jatropha curcas] |
| 3 |
Hb_000723_120 |
0.1187975435 |
- |
- |
PREDICTED: UDP-glycosyltransferase 86A1 [Jatropha curcas] |
| 4 |
Hb_002518_270 |
0.1288910229 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 5 |
Hb_000120_070 |
0.1298498907 |
- |
- |
PREDICTED: uncharacterized protein LOC105630351 [Jatropha curcas] |
| 6 |
Hb_006501_170 |
0.1394235842 |
- |
- |
hypothetical protein JCGZ_07371 [Jatropha curcas] |
| 7 |
Hb_004459_110 |
0.1403599759 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X2 [Populus euphratica] |
| 8 |
Hb_000223_050 |
0.1406029105 |
- |
- |
PREDICTED: endochitinase A-like [Jatropha curcas] |
| 9 |
Hb_000030_260 |
0.141322261 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 10 |
Hb_003556_070 |
0.1455621379 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 11 |
Hb_003337_050 |
0.1484025384 |
- |
- |
PREDICTED: probable protein phosphatase 2C 78 [Jatropha curcas] |
| 12 |
Hb_073119_010 |
0.1494081333 |
- |
- |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 13 |
Hb_004007_120 |
0.1507020782 |
- |
- |
PREDICTED: calcium-transporting ATPase 2, plasma membrane-type isoform X1 [Populus euphratica] |
| 14 |
Hb_004218_160 |
0.1563911828 |
- |
- |
NBS-LRR type disease resistance protein [Medicago truncatula] |
| 15 |
Hb_002085_070 |
0.1571590078 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_000265_020 |
0.1580152452 |
- |
- |
hypothetical protein POPTR_0011s16980g, partial [Populus trichocarpa] |
| 17 |
Hb_000252_080 |
0.1590340882 |
- |
- |
PREDICTED: uncharacterized protein LOC105644644 [Jatropha curcas] |
| 18 |
Hb_000977_090 |
0.1590472985 |
- |
- |
hypothetical protein POPTR_0010s18550g [Populus trichocarpa] |
| 19 |
Hb_000280_110 |
0.1606558016 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 20 |
Hb_000075_110 |
0.1607979896 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |