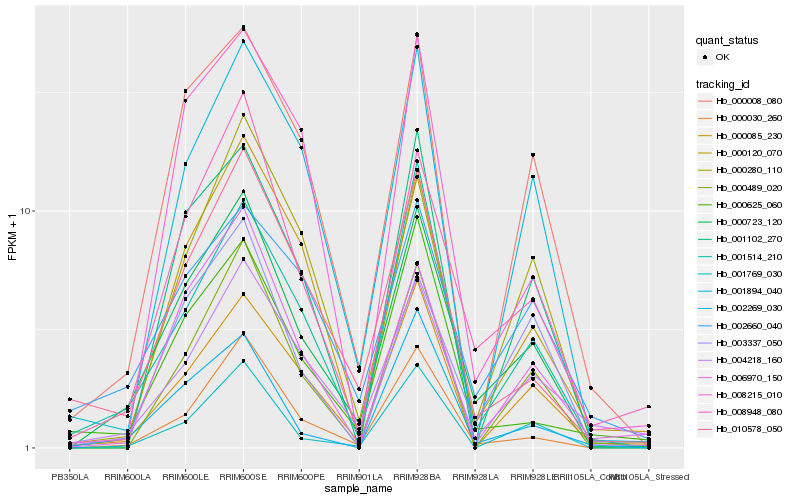
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000723_120 |
0.0 |
- |
- |
PREDICTED: UDP-glycosyltransferase 86A1 [Jatropha curcas] |
| 2 |
Hb_008948_080 |
0.1187975435 |
- |
- |
PREDICTED: TBC1 domain family member 13-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000030_260 |
0.1360889886 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 4 |
Hb_010578_050 |
0.1458610002 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000120_070 |
0.149251418 |
- |
- |
PREDICTED: uncharacterized protein LOC105630351 [Jatropha curcas] |
| 6 |
Hb_000008_080 |
0.1501107202 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001514_210 |
0.1513302544 |
- |
- |
hypothetical protein VITISV_012133 [Vitis vinifera] |
| 8 |
Hb_004218_160 |
0.1513917786 |
- |
- |
NBS-LRR type disease resistance protein [Medicago truncatula] |
| 9 |
Hb_000085_230 |
0.1522005183 |
- |
- |
PREDICTED: probable long-chain-alcohol O-fatty-acyltransferase 5 [Jatropha curcas] |
| 10 |
Hb_001769_030 |
0.1568385952 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 11 |
Hb_006970_150 |
0.1584606161 |
- |
- |
PREDICTED: uncharacterized protein LOC105650471 [Jatropha curcas] |
| 12 |
Hb_000489_020 |
0.1586006824 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Populus euphratica] |
| 13 |
Hb_001894_040 |
0.1615010467 |
- |
- |
PREDICTED: carbonic anhydrase 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003337_050 |
0.162594669 |
- |
- |
PREDICTED: probable protein phosphatase 2C 78 [Jatropha curcas] |
| 15 |
Hb_000280_110 |
0.1647811323 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 16 |
Hb_001102_270 |
0.1660409559 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 17 |
Hb_000625_060 |
0.1661067713 |
- |
- |
Protein P21, putative [Ricinus communis] |
| 18 |
Hb_002660_040 |
0.1670027177 |
- |
- |
PREDICTED: CASP-like protein 5A1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002269_030 |
0.1670135622 |
- |
- |
hypothetical protein CISIN_1g018400mg [Citrus sinensis] |
| 20 |
Hb_008215_010 |
0.1675293675 |
- |
- |
conserved hypothetical protein [Ricinus communis] |