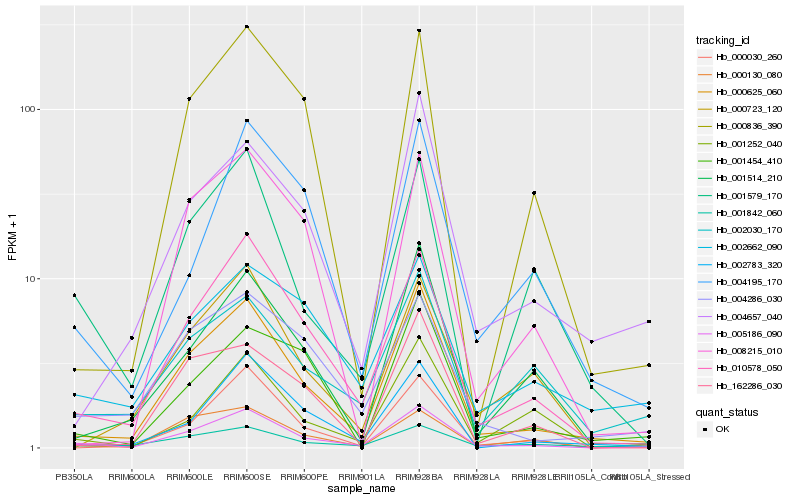
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005186_090 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_000625_060 |
0.1059466714 |
- |
- |
Protein P21, putative [Ricinus communis] |
| 3 |
Hb_010578_050 |
0.112636113 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000030_260 |
0.1645966428 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 5 |
Hb_000130_080 |
0.1650440162 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_004286_030 |
0.1675345737 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 7 |
Hb_001842_060 |
0.1733746874 |
desease resistance |
Gene Name: ABC_trans_N |
hypothetical protein JCGZ_10908 [Jatropha curcas] |
| 8 |
Hb_002783_320 |
0.1775195825 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FC isoform X1 [Jatropha curcas] |
| 9 |
Hb_001514_210 |
0.1821801974 |
- |
- |
hypothetical protein VITISV_012133 [Vitis vinifera] |
| 10 |
Hb_001252_040 |
0.1826600519 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 11 |
Hb_002030_170 |
0.1830066113 |
- |
- |
PREDICTED: calcium-transporting ATPase 1-like [Jatropha curcas] |
| 12 |
Hb_001579_170 |
0.1832658575 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_002662_090 |
0.1881659706 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 14 |
Hb_000723_120 |
0.1883582392 |
- |
- |
PREDICTED: UDP-glycosyltransferase 86A1 [Jatropha curcas] |
| 15 |
Hb_004657_040 |
0.1886810118 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 15 family protein [Populus trichocarpa] |
| 16 |
Hb_162286_030 |
0.1887946335 |
- |
- |
PREDICTED: uncharacterized protein LOC105650163 [Jatropha curcas] |
| 17 |
Hb_000836_390 |
0.1896416742 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 18 |
Hb_004195_170 |
0.1904236608 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_008215_010 |
0.1926727674 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001454_410 |
0.1967113188 |
- |
- |
PREDICTED: uncharacterized protein LOC105643453 [Jatropha curcas] |