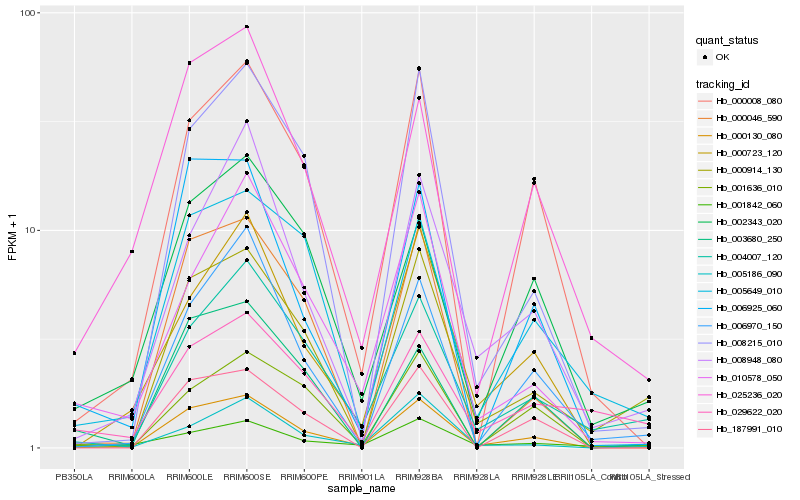
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000130_080 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_000914_130 |
0.1196528164 |
- |
- |
copper-transporting atpase p-type, putative [Ricinus communis] |
| 3 |
Hb_004007_120 |
0.1526213093 |
- |
- |
PREDICTED: calcium-transporting ATPase 2, plasma membrane-type isoform X1 [Populus euphratica] |
| 4 |
Hb_006925_060 |
0.1609182901 |
- |
- |
hypothetical protein JCGZ_20651 [Jatropha curcas] |
| 5 |
Hb_005186_090 |
0.1650440162 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_006970_150 |
0.1653213864 |
- |
- |
PREDICTED: uncharacterized protein LOC105650471 [Jatropha curcas] |
| 7 |
Hb_002343_020 |
0.1654507184 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 8 |
Hb_005649_010 |
0.1663385447 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 9 |
Hb_008215_010 |
0.1663488178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000008_080 |
0.1666374597 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000046_590 |
0.1691254141 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 12 |
Hb_003680_250 |
0.1699017027 |
- |
- |
PREDICTED: probable 3-deoxy-D-manno-octulosonic acid transferase, mitochondrial isoform X2 [Jatropha curcas] |
| 13 |
Hb_001842_060 |
0.1715874728 |
desease resistance |
Gene Name: ABC_trans_N |
hypothetical protein JCGZ_10908 [Jatropha curcas] |
| 14 |
Hb_000723_120 |
0.1719701914 |
- |
- |
PREDICTED: UDP-glycosyltransferase 86A1 [Jatropha curcas] |
| 15 |
Hb_001636_010 |
0.1736133405 |
- |
- |
hypothetical protein JCGZ_01264 [Jatropha curcas] |
| 16 |
Hb_010578_050 |
0.1748945071 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_008948_080 |
0.175407246 |
- |
- |
PREDICTED: TBC1 domain family member 13-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_025236_020 |
0.1773429852 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 19 |
Hb_187991_010 |
0.1773585298 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 20 |
Hb_029622_020 |
0.1774074831 |
- |
- |
conserved hypothetical protein [Ricinus communis] |