| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
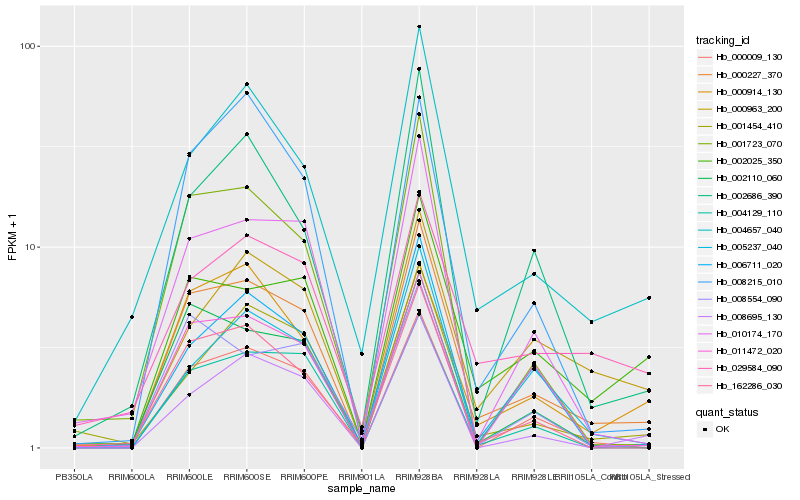
Hb_000227_370 |
0.0 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 2 |
Hb_000009_130 |
0.1259858835 |
- |
- |
hypothetical protein POPTR_0007s12410g [Populus trichocarpa] |
| 3 |
Hb_002025_350 |
0.1415285642 |
- |
- |
PREDICTED: glutamate decarboxylase 4 [Jatropha curcas] |
| 4 |
Hb_162286_030 |
0.1440239774 |
- |
- |
PREDICTED: uncharacterized protein LOC105650163 [Jatropha curcas] |
| 5 |
Hb_002110_060 |
0.145107453 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 6 |
Hb_004657_040 |
0.1464868804 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 15 family protein [Populus trichocarpa] |
| 7 |
Hb_008695_130 |
0.1477178692 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_010174_170 |
0.149013271 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004129_110 |
0.1502606393 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 10 |
Hb_001454_410 |
0.1538186496 |
- |
- |
PREDICTED: uncharacterized protein LOC105643453 [Jatropha curcas] |
| 11 |
Hb_001723_070 |
0.1551576625 |
- |
- |
PREDICTED: polygalacturonase At1g48100 [Jatropha curcas] |
| 12 |
Hb_000914_130 |
0.1554895957 |
- |
- |
copper-transporting atpase p-type, putative [Ricinus communis] |
| 13 |
Hb_002686_390 |
0.1579237072 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |
| 14 |
Hb_006711_020 |
0.1623843506 |
- |
- |
hypothetical protein JCGZ_07258 [Jatropha curcas] |
| 15 |
Hb_000963_200 |
0.1656385077 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 16 |
Hb_005237_040 |
0.1663612286 |
- |
- |
PREDICTED: uncharacterized protein LOC105630923 [Jatropha curcas] |
| 17 |
Hb_008215_010 |
0.167127308 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_008554_090 |
0.1681018319 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_029584_090 |
0.1689621628 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 20 |
Hb_011472_020 |
0.1699058021 |
- |
- |
PREDICTED: mucin-17-like [Jatropha curcas] |