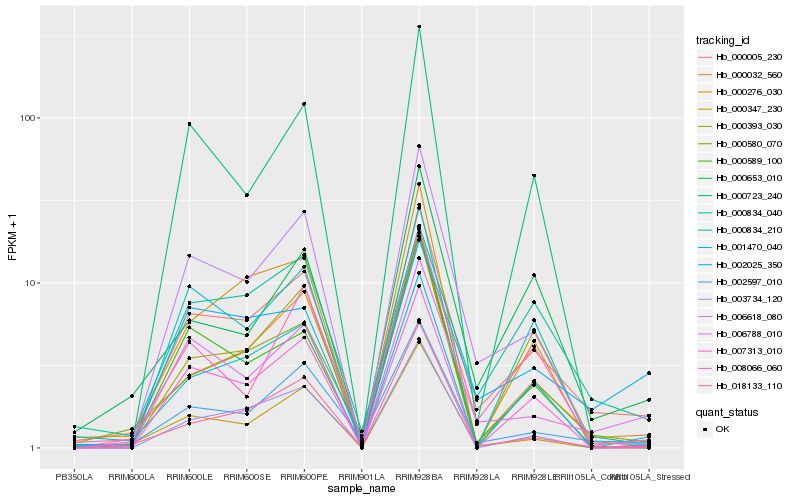
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000005_230 |
0.0 |
- |
- |
hypothetical protein POPTR_0009s01160g [Populus trichocarpa] |
| 2 |
Hb_006788_010 |
0.1151180223 |
- |
- |
PREDICTED: calcium-transporting ATPase 12, plasma membrane-type [Jatropha curcas] |
| 3 |
Hb_002597_010 |
0.1157336775 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Malus domestica] |
| 4 |
Hb_006618_080 |
0.1169551954 |
- |
- |
PREDICTED: non-specific lipid-transfer protein [Jatropha curcas] |
| 5 |
Hb_000032_560 |
0.12814678 |
- |
- |
hypothetical protein JCGZ_15037 [Jatropha curcas] |
| 6 |
Hb_007313_010 |
0.1405001498 |
- |
- |
hypothetical protein JCGZ_23398 [Jatropha curcas] |
| 7 |
Hb_000347_230 |
0.1449995017 |
- |
- |
inter-alpha-trypsin inhibitor heavy chain, putative [Ricinus communis] |
| 8 |
Hb_000580_070 |
0.1456961778 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 9 |
Hb_002025_350 |
0.1476275809 |
- |
- |
PREDICTED: glutamate decarboxylase 4 [Jatropha curcas] |
| 10 |
Hb_000834_210 |
0.1508426684 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_008066_060 |
0.15206678 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 12 |
Hb_018133_110 |
0.1535989033 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 13 |
Hb_001470_040 |
0.1551904659 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 3 [Jatropha curcas] |
| 14 |
Hb_000834_040 |
0.1559818088 |
- |
- |
PREDICTED: ras-related protein Rab7 [Cucumis melo] |
| 15 |
Hb_000589_100 |
0.156317681 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_000723_240 |
0.1574584046 |
- |
- |
phenylalanine ammonia-lyase 2 [Manihot esculenta] |
| 17 |
Hb_003734_120 |
0.1588738678 |
- |
- |
PREDICTED: uncharacterized protein LOC105632812 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000276_030 |
0.1588911265 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 19 |
Hb_000393_030 |
0.1591742269 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 20 |
Hb_000653_010 |
0.1621850368 |
- |
- |
hypothetical protein CICLE_v10033238mg [Citrus clementina] |