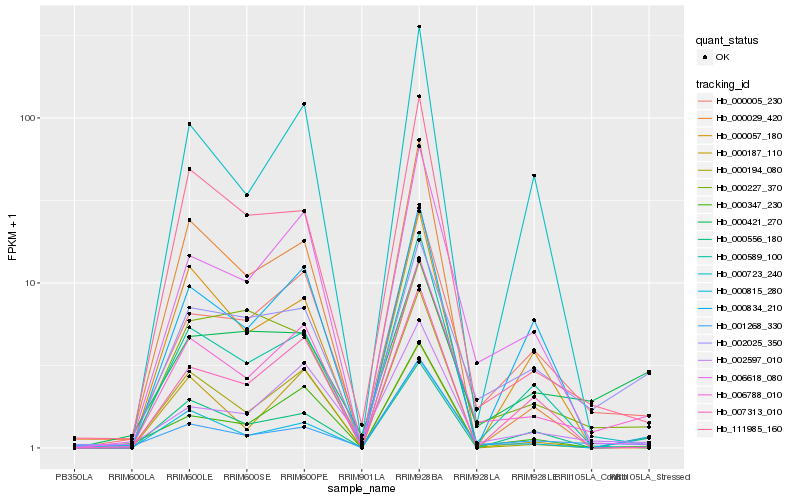
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006788_010 |
0.0 |
- |
- |
PREDICTED: calcium-transporting ATPase 12, plasma membrane-type [Jatropha curcas] |
| 2 |
Hb_000005_230 |
0.1151180223 |
- |
- |
hypothetical protein POPTR_0009s01160g [Populus trichocarpa] |
| 3 |
Hb_006618_080 |
0.1315331757 |
- |
- |
PREDICTED: non-specific lipid-transfer protein [Jatropha curcas] |
| 4 |
Hb_002025_350 |
0.1484235958 |
- |
- |
PREDICTED: glutamate decarboxylase 4 [Jatropha curcas] |
| 5 |
Hb_002597_010 |
0.152653001 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Malus domestica] |
| 6 |
Hb_000347_230 |
0.1555143513 |
- |
- |
inter-alpha-trypsin inhibitor heavy chain, putative [Ricinus communis] |
| 7 |
Hb_000815_280 |
0.1637430257 |
- |
- |
PREDICTED: phosphate transporter PHO1 homolog 3 [Jatropha curcas] |
| 8 |
Hb_000556_180 |
0.1657695593 |
- |
- |
hypothetical protein POPTR_0014s06690g [Populus trichocarpa] |
| 9 |
Hb_000589_100 |
0.1705745324 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_001268_330 |
0.1730161923 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 11 |
Hb_111985_160 |
0.1731809944 |
- |
- |
hypothetical protein B456_010G008900 [Gossypium raimondii] |
| 12 |
Hb_000194_080 |
0.177533105 |
- |
- |
hypothetical protein JCGZ_18743 [Jatropha curcas] |
| 13 |
Hb_000723_240 |
0.1801978359 |
- |
- |
phenylalanine ammonia-lyase 2 [Manihot esculenta] |
| 14 |
Hb_007313_010 |
0.1831614561 |
- |
- |
hypothetical protein JCGZ_23398 [Jatropha curcas] |
| 15 |
Hb_000057_180 |
0.1847137827 |
- |
- |
hypothetical protein POPTR_0001s18200g [Populus trichocarpa] |
| 16 |
Hb_000029_420 |
0.1874701833 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 17 |
Hb_000834_210 |
0.1874972145 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_000421_270 |
0.1875530424 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 1 [Theobroma cacao] |
| 19 |
Hb_000227_370 |
0.1881694008 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 20 |
Hb_000187_110 |
0.1898542594 |
- |
- |
PREDICTED: serine carboxypeptidase-like 7 [Jatropha curcas] |