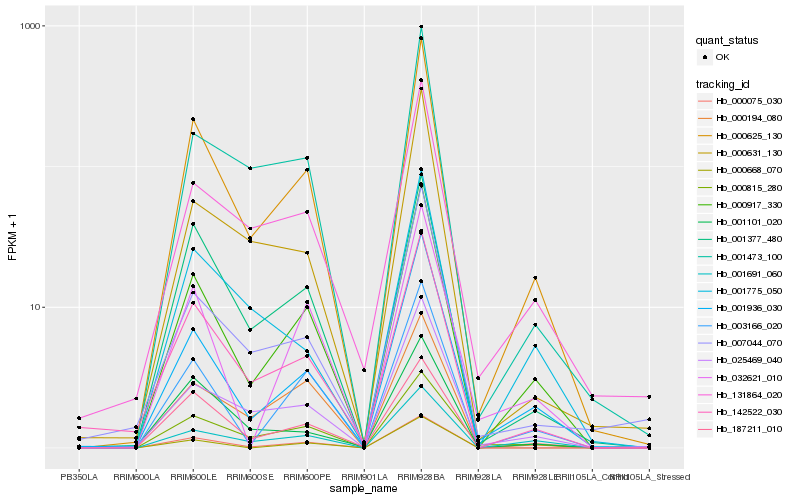
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000625_130 |
0.0 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase [Jatropha curcas] |
| 2 |
Hb_000917_330 |
0.0520554726 |
- |
- |
PREDICTED: leucoanthocyanidin reductase-like [Jatropha curcas] |
| 3 |
Hb_001936_030 |
0.0893002081 |
- |
- |
PREDICTED: polygalacturonase At1g48100 isoform X1 [Jatropha curcas] |
| 4 |
Hb_142522_030 |
0.1000790939 |
- |
- |
hypothetical protein JCGZ_21596 [Jatropha curcas] |
| 5 |
Hb_001377_480 |
0.1012170915 |
transcription factor |
TF Family: MYB |
Myb domain protein 7, putative [Theobroma cacao] |
| 6 |
Hb_025469_040 |
0.1015354564 |
- |
- |
PREDICTED: acidic mammalian chitinase-like [Jatropha curcas] |
| 7 |
Hb_003166_020 |
0.1143537666 |
- |
- |
PREDICTED: RNA-binding E3 ubiquitin-protein ligase MEX3C-like [Jatropha curcas] |
| 8 |
Hb_001691_060 |
0.115773279 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 9 |
Hb_001473_100 |
0.1168131997 |
- |
- |
chalcone synthase 1 [Jatropha curcas] |
| 10 |
Hb_000815_280 |
0.1181200212 |
- |
- |
PREDICTED: phosphate transporter PHO1 homolog 3 [Jatropha curcas] |
| 11 |
Hb_131864_020 |
0.1220774672 |
- |
- |
PREDICTED: anthocyanidin reductase [Jatropha curcas] |
| 12 |
Hb_187211_010 |
0.1275591402 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 13 |
Hb_001101_020 |
0.1306567711 |
- |
- |
hypothetical protein RCOM_0615680 [Ricinus communis] |
| 14 |
Hb_007044_070 |
0.1312115722 |
- |
- |
PREDICTED: tetraketide alpha-pyrone reductase 1-like [Jatropha curcas] |
| 15 |
Hb_000631_130 |
0.1319880613 |
- |
- |
PREDICTED: flavonoid 3'-monooxygenase [Jatropha curcas] |
| 16 |
Hb_000075_030 |
0.1328866497 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C10-like [Fragaria vesca subsp. vesca] |
| 17 |
Hb_000194_080 |
0.1346845783 |
- |
- |
hypothetical protein JCGZ_18743 [Jatropha curcas] |
| 18 |
Hb_001775_050 |
0.1383320573 |
- |
- |
ribosome-inactivating protein cucurmosin-like [Jatropha curcas] |
| 19 |
Hb_032621_010 |
0.1414035176 |
- |
- |
hypothetical protein PHAVU_006G197300g [Phaseolus vulgaris] |
| 20 |
Hb_000668_070 |
0.1469674567 |
- |
- |
conserved hypothetical protein [Ricinus communis] |