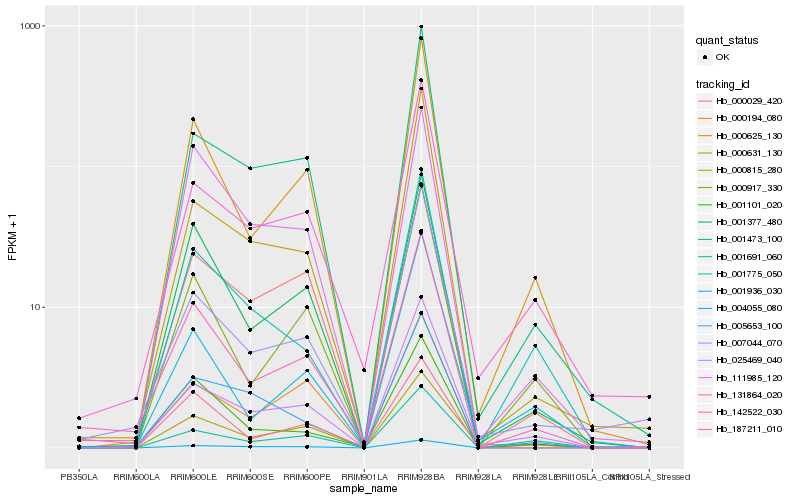
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_142522_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_21596 [Jatropha curcas] |
| 2 |
Hb_000625_130 |
0.1000790939 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase [Jatropha curcas] |
| 3 |
Hb_001101_020 |
0.1145777427 |
- |
- |
hypothetical protein RCOM_0615680 [Ricinus communis] |
| 4 |
Hb_001377_480 |
0.1188937323 |
transcription factor |
TF Family: MYB |
Myb domain protein 7, putative [Theobroma cacao] |
| 5 |
Hb_000917_330 |
0.1261947271 |
- |
- |
PREDICTED: leucoanthocyanidin reductase-like [Jatropha curcas] |
| 6 |
Hb_131864_020 |
0.1265484025 |
- |
- |
PREDICTED: anthocyanidin reductase [Jatropha curcas] |
| 7 |
Hb_007044_070 |
0.1290196752 |
- |
- |
PREDICTED: tetraketide alpha-pyrone reductase 1-like [Jatropha curcas] |
| 8 |
Hb_025469_040 |
0.1291540065 |
- |
- |
PREDICTED: acidic mammalian chitinase-like [Jatropha curcas] |
| 9 |
Hb_187211_010 |
0.1322524226 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 10 |
Hb_000815_280 |
0.1336928098 |
- |
- |
PREDICTED: phosphate transporter PHO1 homolog 3 [Jatropha curcas] |
| 11 |
Hb_001473_100 |
0.1360012689 |
- |
- |
chalcone synthase 1 [Jatropha curcas] |
| 12 |
Hb_000631_130 |
0.1385282387 |
- |
- |
PREDICTED: flavonoid 3'-monooxygenase [Jatropha curcas] |
| 13 |
Hb_001936_030 |
0.1410635592 |
- |
- |
PREDICTED: polygalacturonase At1g48100 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000029_420 |
0.1425053224 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 15 |
Hb_004055_080 |
0.1511072906 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 16 |
Hb_000194_080 |
0.1521806509 |
- |
- |
hypothetical protein JCGZ_18743 [Jatropha curcas] |
| 17 |
Hb_001775_050 |
0.1524390001 |
- |
- |
ribosome-inactivating protein cucurmosin-like [Jatropha curcas] |
| 18 |
Hb_111985_120 |
0.1546732215 |
- |
- |
dihydroflavonol-4-reductase [Vitis rotundifolia] |
| 19 |
Hb_005653_100 |
0.1571844742 |
- |
- |
hypothetical protein POPTR_0004s19580g [Populus trichocarpa] |
| 20 |
Hb_001691_060 |
0.160932081 |
- |
- |
lipoxygenase, putative [Ricinus communis] |