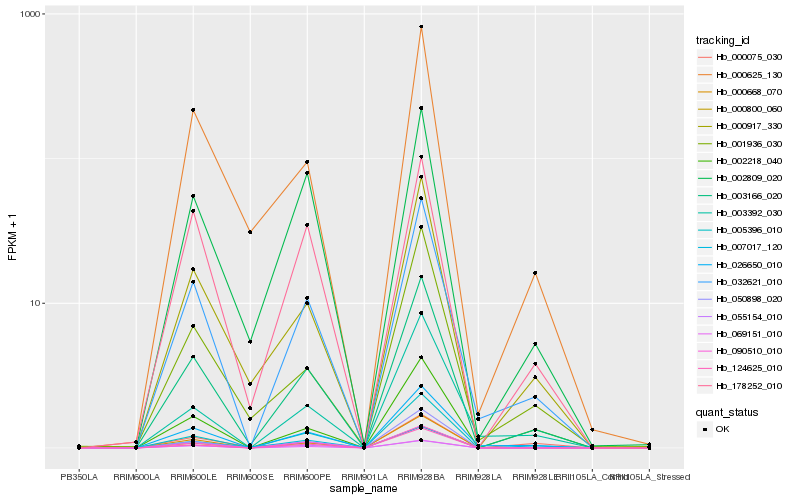
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003166_020 |
0.0 |
- |
- |
PREDICTED: RNA-binding E3 ubiquitin-protein ligase MEX3C-like [Jatropha curcas] |
| 2 |
Hb_032621_010 |
0.0716296966 |
- |
- |
hypothetical protein PHAVU_006G197300g [Phaseolus vulgaris] |
| 3 |
Hb_000917_330 |
0.0864665151 |
- |
- |
PREDICTED: leucoanthocyanidin reductase-like [Jatropha curcas] |
| 4 |
Hb_026650_010 |
0.0930507453 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 5 |
Hb_000800_060 |
0.103803493 |
- |
- |
hypothetical protein POPTR_0008s02870g [Populus trichocarpa] |
| 6 |
Hb_000668_070 |
0.1068185112 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_069151_010 |
0.1071257891 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Gossypium raimondii] |
| 8 |
Hb_090510_010 |
0.1085571311 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 9 |
Hb_000625_130 |
0.1143537666 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase [Jatropha curcas] |
| 10 |
Hb_005396_010 |
0.1152995202 |
- |
- |
PREDICTED: interleukin enhancer-binding factor 3-B-like [Jatropha curcas] |
| 11 |
Hb_002809_020 |
0.1207493047 |
- |
- |
PREDICTED: leucoanthocyanidin dioxygenase [Jatropha curcas] |
| 12 |
Hb_124625_010 |
0.1210343591 |
- |
- |
hypothetical protein JCGZ_01142 [Jatropha curcas] |
| 13 |
Hb_178252_010 |
0.1241027017 |
- |
- |
hypothetical protein POPTR_0018s01890g [Populus trichocarpa] |
| 14 |
Hb_007017_120 |
0.1260480638 |
- |
- |
PREDICTED: exocyst complex component EXO84A [Jatropha curcas] |
| 15 |
Hb_001936_030 |
0.1277852036 |
- |
- |
PREDICTED: polygalacturonase At1g48100 isoform X1 [Jatropha curcas] |
| 16 |
Hb_050898_020 |
0.1343826826 |
- |
- |
hypothetical protein JCGZ_19965 [Jatropha curcas] |
| 17 |
Hb_002218_040 |
0.1386453308 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000075_030 |
0.139734395 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C10-like [Fragaria vesca subsp. vesca] |
| 19 |
Hb_055154_010 |
0.1435174556 |
- |
- |
PREDICTED: laccase-7 [Jatropha curcas] |
| 20 |
Hb_003392_030 |
0.1435629728 |
- |
- |
conserved hypothetical protein [Ricinus communis] |