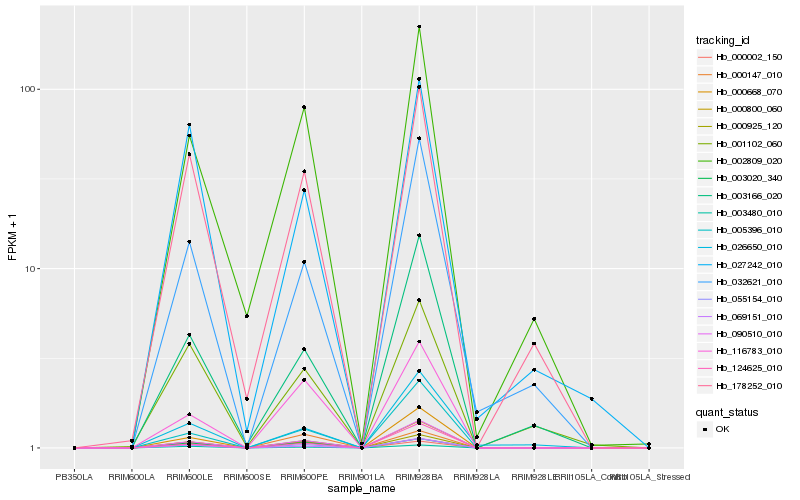
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_055154_010 |
0.0 |
- |
- |
PREDICTED: laccase-7 [Jatropha curcas] |
| 2 |
Hb_000925_120 |
0.0782370582 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 3 |
Hb_090510_010 |
0.0873171255 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 4 |
Hb_000800_060 |
0.0990176394 |
- |
- |
hypothetical protein POPTR_0008s02870g [Populus trichocarpa] |
| 5 |
Hb_069151_010 |
0.1050043484 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Gossypium raimondii] |
| 6 |
Hb_003020_340 |
0.1050683204 |
- |
- |
PREDICTED: uncharacterized protein LOC105631327 [Jatropha curcas] |
| 7 |
Hb_116783_010 |
0.1082189761 |
- |
- |
hypothetical protein POPTR_0011s16190g [Populus trichocarpa] |
| 8 |
Hb_178252_010 |
0.1151120466 |
- |
- |
hypothetical protein POPTR_0018s01890g [Populus trichocarpa] |
| 9 |
Hb_003480_010 |
0.1170561896 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 10 |
Hb_002809_020 |
0.1183272499 |
- |
- |
PREDICTED: leucoanthocyanidin dioxygenase [Jatropha curcas] |
| 11 |
Hb_005396_010 |
0.1253787565 |
- |
- |
PREDICTED: interleukin enhancer-binding factor 3-B-like [Jatropha curcas] |
| 12 |
Hb_003166_020 |
0.1435174556 |
- |
- |
PREDICTED: RNA-binding E3 ubiquitin-protein ligase MEX3C-like [Jatropha curcas] |
| 13 |
Hb_032621_010 |
0.1449500001 |
- |
- |
hypothetical protein PHAVU_006G197300g [Phaseolus vulgaris] |
| 14 |
Hb_000002_150 |
0.1469671281 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000147_010 |
0.1485260857 |
- |
- |
PREDICTED: UDP-glycosyltransferase 87A1-like [Jatropha curcas] |
| 16 |
Hb_000668_070 |
0.1511280275 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_124625_010 |
0.1511373004 |
- |
- |
hypothetical protein JCGZ_01142 [Jatropha curcas] |
| 18 |
Hb_027242_010 |
0.1612021276 |
- |
- |
PREDICTED: pathogenesis-related protein 1-like [Populus euphratica] |
| 19 |
Hb_001102_060 |
0.1635120732 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At1g78530 isoform X1 [Jatropha curcas] |
| 20 |
Hb_026650_010 |
0.1690707901 |
- |
- |
nitrate transporter, putative [Ricinus communis] |