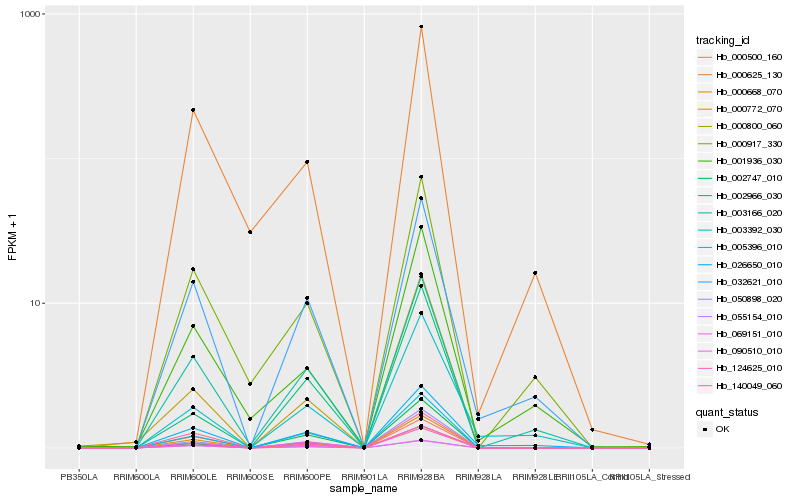
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000668_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_069151_010 |
0.0674970342 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Gossypium raimondii] |
| 3 |
Hb_000800_060 |
0.0730255161 |
- |
- |
hypothetical protein POPTR_0008s02870g [Populus trichocarpa] |
| 4 |
Hb_050898_020 |
0.0734840419 |
- |
- |
hypothetical protein JCGZ_19965 [Jatropha curcas] |
| 5 |
Hb_124625_010 |
0.076570398 |
- |
- |
hypothetical protein JCGZ_01142 [Jatropha curcas] |
| 6 |
Hb_005396_010 |
0.082891451 |
- |
- |
PREDICTED: interleukin enhancer-binding factor 3-B-like [Jatropha curcas] |
| 7 |
Hb_090510_010 |
0.0880832183 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 8 |
Hb_003166_020 |
0.1068185112 |
- |
- |
PREDICTED: RNA-binding E3 ubiquitin-protein ligase MEX3C-like [Jatropha curcas] |
| 9 |
Hb_140049_060 |
0.1097121782 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 10 |
Hb_000772_070 |
0.1204281855 |
- |
- |
PREDICTED: uncharacterized protein LOC105649697 [Jatropha curcas] |
| 11 |
Hb_032621_010 |
0.1234081157 |
- |
- |
hypothetical protein PHAVU_006G197300g [Phaseolus vulgaris] |
| 12 |
Hb_026650_010 |
0.1324905053 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 13 |
Hb_000917_330 |
0.1383295733 |
- |
- |
PREDICTED: leucoanthocyanidin reductase-like [Jatropha curcas] |
| 14 |
Hb_000625_130 |
0.1469674567 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase [Jatropha curcas] |
| 15 |
Hb_055154_010 |
0.1511280275 |
- |
- |
PREDICTED: laccase-7 [Jatropha curcas] |
| 16 |
Hb_000500_160 |
0.1532010359 |
- |
- |
hypothetical protein CISIN_1g039640mg, partial [Citrus sinensis] |
| 17 |
Hb_001936_030 |
0.1551844807 |
- |
- |
PREDICTED: polygalacturonase At1g48100 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002966_030 |
0.1597504574 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002747_010 |
0.1665167733 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 1 [Jatropha curcas] |
| 20 |
Hb_003392_030 |
0.1669567031 |
- |
- |
conserved hypothetical protein [Ricinus communis] |