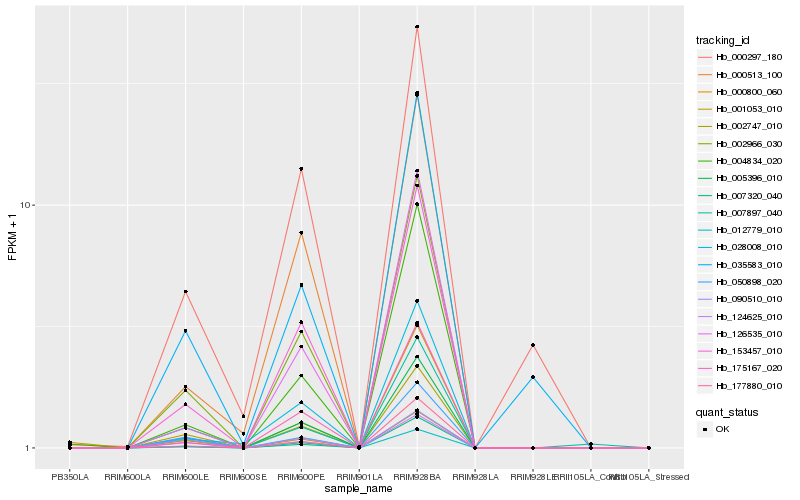
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002747_010 |
0.0 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 1 [Jatropha curcas] |
| 2 |
Hb_002966_030 |
0.0254232797 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_153457_010 |
0.0266833588 |
- |
- |
Miraculin precursor, putative [Ricinus communis] |
| 4 |
Hb_012779_010 |
0.0470739264 |
- |
- |
hypothetical protein JCGZ_05865 [Jatropha curcas] |
| 5 |
Hb_175167_020 |
0.0704461176 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000513_100 |
0.0837628455 |
- |
- |
germin-like protein GLP, partial [Manihot esculenta] |
| 7 |
Hb_007320_040 |
0.0912832383 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |
| 8 |
Hb_124625_010 |
0.0927509624 |
- |
- |
hypothetical protein JCGZ_01142 [Jatropha curcas] |
| 9 |
Hb_028008_010 |
0.092859036 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 10 |
Hb_005396_010 |
0.1014284872 |
- |
- |
PREDICTED: interleukin enhancer-binding factor 3-B-like [Jatropha curcas] |
| 11 |
Hb_177880_010 |
0.1033283748 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 12 |
Hb_050898_020 |
0.1060915668 |
- |
- |
hypothetical protein JCGZ_19965 [Jatropha curcas] |
| 13 |
Hb_126535_010 |
0.1074006147 |
- |
- |
hypothetical protein JCGZ_09554 [Jatropha curcas] |
| 14 |
Hb_004834_020 |
0.1087915558 |
- |
- |
major allergen Pru ar 1-like [Jatropha curcas] |
| 15 |
Hb_007897_040 |
0.1167461939 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_001053_010 |
0.1202810634 |
- |
- |
PREDICTED: polygalacturonase-like [Jatropha curcas] |
| 17 |
Hb_000297_180 |
0.1255183342 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 18 |
Hb_000800_060 |
0.1323786921 |
- |
- |
hypothetical protein POPTR_0008s02870g [Populus trichocarpa] |
| 19 |
Hb_090510_010 |
0.1340024047 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 20 |
Hb_035583_010 |
0.1341258053 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |