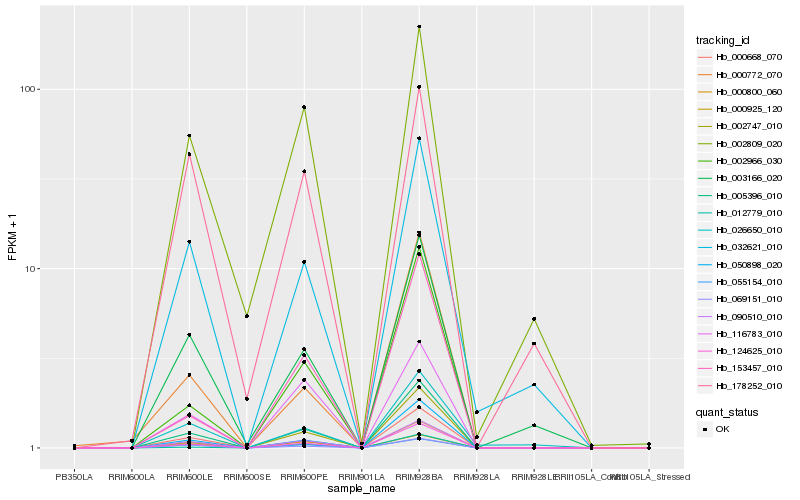
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_090510_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 2 |
Hb_000800_060 |
0.0158040048 |
- |
- |
hypothetical protein POPTR_0008s02870g [Populus trichocarpa] |
| 3 |
Hb_005396_010 |
0.0386230867 |
- |
- |
PREDICTED: interleukin enhancer-binding factor 3-B-like [Jatropha curcas] |
| 4 |
Hb_124625_010 |
0.0642520747 |
- |
- |
hypothetical protein JCGZ_01142 [Jatropha curcas] |
| 5 |
Hb_069151_010 |
0.0867634522 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Gossypium raimondii] |
| 6 |
Hb_055154_010 |
0.0873171255 |
- |
- |
PREDICTED: laccase-7 [Jatropha curcas] |
| 7 |
Hb_000668_070 |
0.0880832183 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_050898_020 |
0.1025940147 |
- |
- |
hypothetical protein JCGZ_19965 [Jatropha curcas] |
| 9 |
Hb_003166_020 |
0.1085571311 |
- |
- |
PREDICTED: RNA-binding E3 ubiquitin-protein ligase MEX3C-like [Jatropha curcas] |
| 10 |
Hb_032621_010 |
0.1217471507 |
- |
- |
hypothetical protein PHAVU_006G197300g [Phaseolus vulgaris] |
| 11 |
Hb_116783_010 |
0.1278905232 |
- |
- |
hypothetical protein POPTR_0011s16190g [Populus trichocarpa] |
| 12 |
Hb_002809_020 |
0.1314786107 |
- |
- |
PREDICTED: leucoanthocyanidin dioxygenase [Jatropha curcas] |
| 13 |
Hb_002747_010 |
0.1340024047 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 1 [Jatropha curcas] |
| 14 |
Hb_026650_010 |
0.1364995526 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 15 |
Hb_000925_120 |
0.140021177 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 16 |
Hb_002966_030 |
0.1414742008 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_012779_010 |
0.1503713972 |
- |
- |
hypothetical protein JCGZ_05865 [Jatropha curcas] |
| 18 |
Hb_153457_010 |
0.1564477498 |
- |
- |
Miraculin precursor, putative [Ricinus communis] |
| 19 |
Hb_178252_010 |
0.1599243271 |
- |
- |
hypothetical protein POPTR_0018s01890g [Populus trichocarpa] |
| 20 |
Hb_000772_070 |
0.1653692445 |
- |
- |
PREDICTED: uncharacterized protein LOC105649697 [Jatropha curcas] |