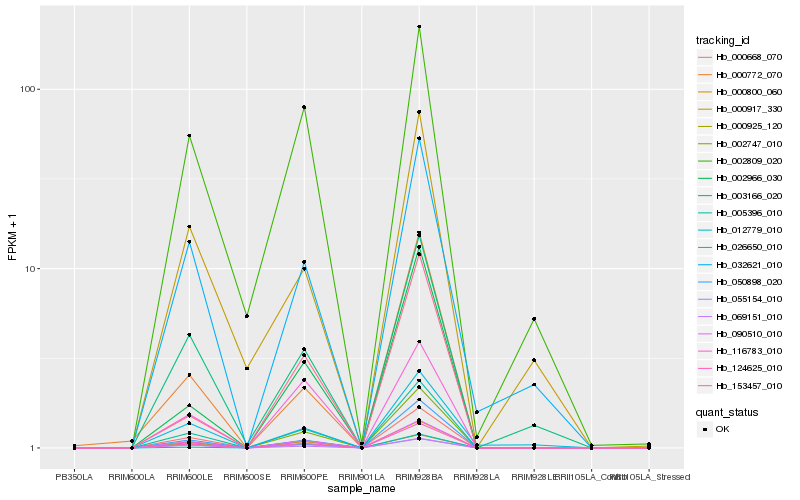
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000800_060 |
0.0 |
- |
- |
hypothetical protein POPTR_0008s02870g [Populus trichocarpa] |
| 2 |
Hb_090510_010 |
0.0158040048 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 3 |
Hb_005396_010 |
0.0321800145 |
- |
- |
PREDICTED: interleukin enhancer-binding factor 3-B-like [Jatropha curcas] |
| 4 |
Hb_124625_010 |
0.0538093599 |
- |
- |
hypothetical protein JCGZ_01142 [Jatropha curcas] |
| 5 |
Hb_000668_070 |
0.0730255161 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_069151_010 |
0.080104324 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Gossypium raimondii] |
| 7 |
Hb_050898_020 |
0.0893162886 |
- |
- |
hypothetical protein JCGZ_19965 [Jatropha curcas] |
| 8 |
Hb_055154_010 |
0.0990176394 |
- |
- |
PREDICTED: laccase-7 [Jatropha curcas] |
| 9 |
Hb_003166_020 |
0.103803493 |
- |
- |
PREDICTED: RNA-binding E3 ubiquitin-protein ligase MEX3C-like [Jatropha curcas] |
| 10 |
Hb_032621_010 |
0.1186133197 |
- |
- |
hypothetical protein PHAVU_006G197300g [Phaseolus vulgaris] |
| 11 |
Hb_026650_010 |
0.1321435818 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 12 |
Hb_002747_010 |
0.1323786921 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 1 [Jatropha curcas] |
| 13 |
Hb_002966_030 |
0.1370435655 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002809_020 |
0.140005223 |
- |
- |
PREDICTED: leucoanthocyanidin dioxygenase [Jatropha curcas] |
| 15 |
Hb_116783_010 |
0.1432021468 |
- |
- |
hypothetical protein POPTR_0011s16190g [Populus trichocarpa] |
| 16 |
Hb_000772_070 |
0.1525892172 |
- |
- |
PREDICTED: uncharacterized protein LOC105649697 [Jatropha curcas] |
| 17 |
Hb_012779_010 |
0.1534837061 |
- |
- |
hypothetical protein JCGZ_05865 [Jatropha curcas] |
| 18 |
Hb_000925_120 |
0.1551116951 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 19 |
Hb_153457_010 |
0.1560170674 |
- |
- |
Miraculin precursor, putative [Ricinus communis] |
| 20 |
Hb_000917_330 |
0.158876233 |
- |
- |
PREDICTED: leucoanthocyanidin reductase-like [Jatropha curcas] |