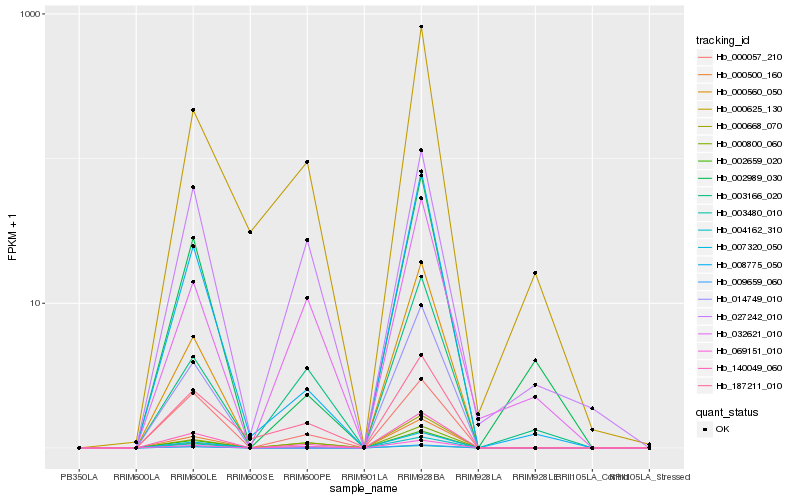
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_140049_060 |
0.0 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 2 |
Hb_000500_160 |
0.0642138262 |
- |
- |
hypothetical protein CISIN_1g039640mg, partial [Citrus sinensis] |
| 3 |
Hb_069151_010 |
0.1051646825 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Gossypium raimondii] |
| 4 |
Hb_008775_050 |
0.1071560541 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Jatropha curcas] |
| 5 |
Hb_000668_070 |
0.1097121782 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000057_210 |
0.1358146407 |
- |
- |
hypothetical protein JCGZ_06274 [Jatropha curcas] |
| 7 |
Hb_187211_010 |
0.1447269075 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 8 |
Hb_002989_030 |
0.1540838089 |
- |
- |
PREDICTED: protein GAST1-like [Populus euphratica] |
| 9 |
Hb_000625_130 |
0.1586929897 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase [Jatropha curcas] |
| 10 |
Hb_007320_050 |
0.1634889769 |
- |
- |
PREDICTED: uncharacterized protein LOC105637288 [Jatropha curcas] |
| 11 |
Hb_004162_310 |
0.1635533021 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 12 |
Hb_014749_010 |
0.1637263025 |
- |
- |
hypothetical protein JCGZ_26951 [Jatropha curcas] |
| 13 |
Hb_003166_020 |
0.1639124352 |
- |
- |
PREDICTED: RNA-binding E3 ubiquitin-protein ligase MEX3C-like [Jatropha curcas] |
| 14 |
Hb_003480_010 |
0.1645264335 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 15 |
Hb_002659_020 |
0.1661281792 |
- |
- |
PREDICTED: cytochrome P450 714C2-like [Populus euphratica] |
| 16 |
Hb_009659_060 |
0.1664959154 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 17 |
Hb_027242_010 |
0.1686498635 |
- |
- |
PREDICTED: pathogenesis-related protein 1-like [Populus euphratica] |
| 18 |
Hb_000800_060 |
0.1698631838 |
- |
- |
hypothetical protein POPTR_0008s02870g [Populus trichocarpa] |
| 19 |
Hb_032621_010 |
0.1703298506 |
- |
- |
hypothetical protein PHAVU_006G197300g [Phaseolus vulgaris] |
| 20 |
Hb_000560_050 |
0.170479955 |
- |
- |
hypothetical protein JCGZ_05738 [Jatropha curcas] |