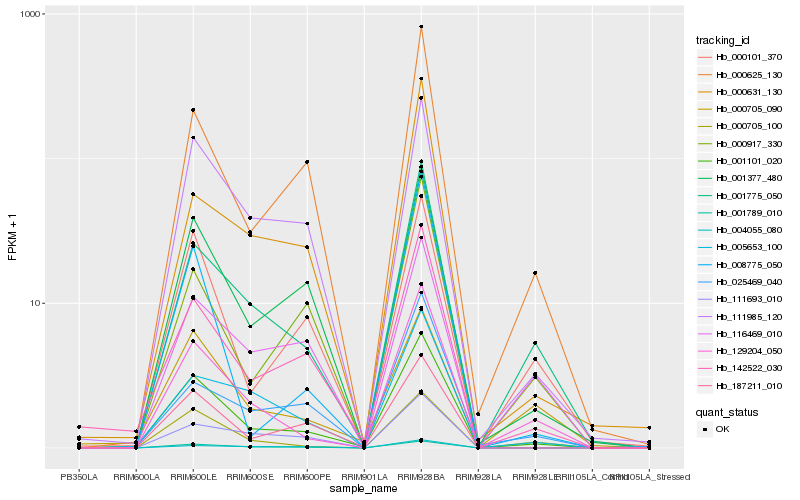
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001101_020 |
0.0 |
- |
- |
hypothetical protein RCOM_0615680 [Ricinus communis] |
| 2 |
Hb_001377_480 |
0.1073086496 |
transcription factor |
TF Family: MYB |
Myb domain protein 7, putative [Theobroma cacao] |
| 3 |
Hb_142522_030 |
0.1145777427 |
- |
- |
hypothetical protein JCGZ_21596 [Jatropha curcas] |
| 4 |
Hb_111985_120 |
0.1177234759 |
- |
- |
dihydroflavonol-4-reductase [Vitis rotundifolia] |
| 5 |
Hb_001789_010 |
0.1200718334 |
- |
- |
hypothetical protein POPTR_0015s13020g [Populus trichocarpa] |
| 6 |
Hb_187211_010 |
0.1209306986 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 7 |
Hb_129204_050 |
0.1224193026 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein 330-like [Prunus mume] |
| 8 |
Hb_001775_050 |
0.1257085471 |
- |
- |
ribosome-inactivating protein cucurmosin-like [Jatropha curcas] |
| 9 |
Hb_000625_130 |
0.1306567711 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase [Jatropha curcas] |
| 10 |
Hb_000705_100 |
0.1431484364 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 11 |
Hb_005653_100 |
0.1451453519 |
- |
- |
hypothetical protein POPTR_0004s19580g [Populus trichocarpa] |
| 12 |
Hb_000101_370 |
0.1466940531 |
- |
- |
PREDICTED: dihydrofolate reductase-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_111693_010 |
0.165361156 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 14 |
Hb_000917_330 |
0.168581442 |
- |
- |
PREDICTED: leucoanthocyanidin reductase-like [Jatropha curcas] |
| 15 |
Hb_116469_010 |
0.169577153 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |
| 16 |
Hb_004055_080 |
0.1698331977 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 17 |
Hb_008775_050 |
0.1699397443 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Jatropha curcas] |
| 18 |
Hb_000705_090 |
0.171038108 |
- |
- |
PREDICTED: uncharacterized protein LOC100246830 [Vitis vinifera] |
| 19 |
Hb_025469_040 |
0.1712051565 |
- |
- |
PREDICTED: acidic mammalian chitinase-like [Jatropha curcas] |
| 20 |
Hb_000631_130 |
0.1728856298 |
- |
- |
PREDICTED: flavonoid 3'-monooxygenase [Jatropha curcas] |