| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
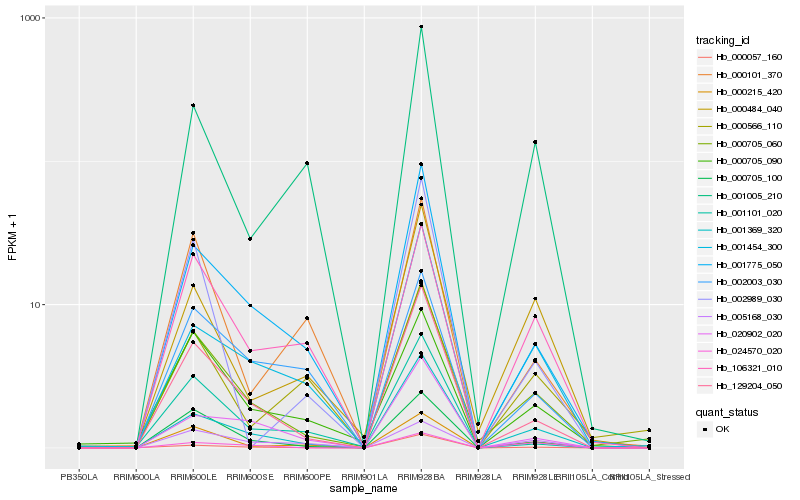
Hb_000705_060 |
0.0 |
- |
- |
hypothetical protein TRIUR3_06087 [Triticum urartu] |
| 2 |
Hb_129204_050 |
0.1187052592 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein 330-like [Prunus mume] |
| 3 |
Hb_000705_100 |
0.1221951984 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 4 |
Hb_000215_420 |
0.1412484551 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2-like [Populus euphratica] |
| 5 |
Hb_001775_050 |
0.146342528 |
- |
- |
ribosome-inactivating protein cucurmosin-like [Jatropha curcas] |
| 6 |
Hb_001369_320 |
0.146783482 |
- |
- |
PREDICTED: dynein light chain, cytoplasmic-like [Jatropha curcas] |
| 7 |
Hb_000705_090 |
0.1579264614 |
- |
- |
PREDICTED: uncharacterized protein LOC100246830 [Vitis vinifera] |
| 8 |
Hb_106321_010 |
0.1723783628 |
- |
- |
PREDICTED: probable glutathione S-transferase [Jatropha curcas] |
| 9 |
Hb_000484_040 |
0.1735933746 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_001454_300 |
0.1772683675 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 11 |
Hb_001101_020 |
0.1835298841 |
- |
- |
hypothetical protein RCOM_0615680 [Ricinus communis] |
| 12 |
Hb_000057_160 |
0.1866421557 |
- |
- |
hypothetical protein JCGZ_06279 [Jatropha curcas] |
| 13 |
Hb_001005_210 |
0.1914954483 |
- |
- |
PREDICTED: chalcone synthase 3 [Jatropha curcas] |
| 14 |
Hb_020902_020 |
0.192116598 |
- |
- |
hypothetical protein JCGZ_07820 [Jatropha curcas] |
| 15 |
Hb_005168_030 |
0.1964222941 |
- |
- |
unknown [Zea mays] |
| 16 |
Hb_002003_030 |
0.1988038637 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 17 |
Hb_000566_110 |
0.1991110693 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Prunus mume] |
| 18 |
Hb_024570_020 |
0.1993838154 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2-like [Populus euphratica] |
| 19 |
Hb_000101_370 |
0.2019818913 |
- |
- |
PREDICTED: dihydrofolate reductase-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_002989_030 |
0.2023091539 |
- |
- |
PREDICTED: protein GAST1-like [Populus euphratica] |