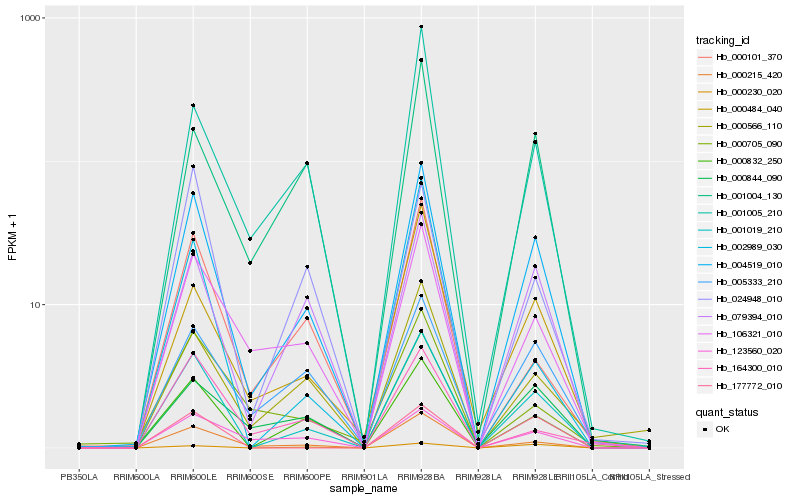
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001019_210 |
0.0 |
- |
- |
PREDICTED: protein kinase PINOID 2 [Gossypium raimondii] |
| 2 |
Hb_004519_010 |
0.0744295554 |
- |
- |
PREDICTED: basic form of pathogenesis-related protein 1-like [Jatropha curcas] |
| 3 |
Hb_000832_250 |
0.1248093659 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC-like [Jatropha curcas] |
| 4 |
Hb_000215_420 |
0.1373170499 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2-like [Populus euphratica] |
| 5 |
Hb_079394_010 |
0.1660578178 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 6 |
Hb_106321_010 |
0.1823865896 |
- |
- |
PREDICTED: probable glutathione S-transferase [Jatropha curcas] |
| 7 |
Hb_000566_110 |
0.1835330025 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Prunus mume] |
| 8 |
Hb_000484_040 |
0.1939871874 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_177772_010 |
0.1942089621 |
- |
- |
PREDICTED: receptor-like protein 12 isoform X2 [Vitis vinifera] |
| 10 |
Hb_000844_090 |
0.194702864 |
- |
- |
PREDICTED: cytochrome P450 82C4-like [Jatropha curcas] |
| 11 |
Hb_024948_010 |
0.1948010083 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase, basic isoform-like [Populus euphratica] |
| 12 |
Hb_001005_210 |
0.1976517978 |
- |
- |
PREDICTED: chalcone synthase 3 [Jatropha curcas] |
| 13 |
Hb_000101_370 |
0.1995263504 |
- |
- |
PREDICTED: dihydrofolate reductase-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_005333_210 |
0.2041305255 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Jatropha curcas] |
| 15 |
Hb_001004_130 |
0.2044154089 |
- |
- |
PREDICTED: chalcone synthase 3 [Jatropha curcas] |
| 16 |
Hb_123560_020 |
0.2129157924 |
- |
- |
hypothetical protein JCGZ_12464 [Jatropha curcas] |
| 17 |
Hb_002989_030 |
0.2149857204 |
- |
- |
PREDICTED: protein GAST1-like [Populus euphratica] |
| 18 |
Hb_000705_090 |
0.2159546722 |
- |
- |
PREDICTED: uncharacterized protein LOC100246830 [Vitis vinifera] |
| 19 |
Hb_000230_020 |
0.2181381644 |
- |
- |
cysteine synthase, putative [Ricinus communis] |
| 20 |
Hb_164300_010 |
0.2195125962 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |