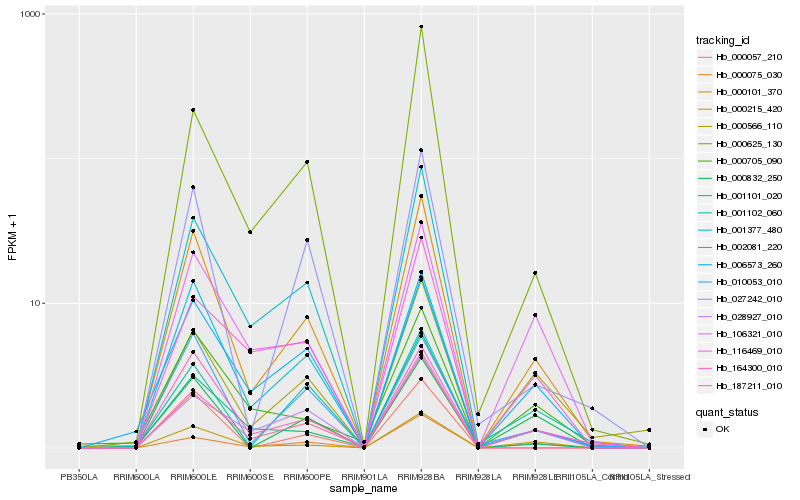
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000101_370 |
0.0 |
- |
- |
PREDICTED: dihydrofolate reductase-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_164300_010 |
0.1098562486 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 3 |
Hb_000215_420 |
0.1145596765 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2-like [Populus euphratica] |
| 4 |
Hb_001377_480 |
0.1253713117 |
transcription factor |
TF Family: MYB |
Myb domain protein 7, putative [Theobroma cacao] |
| 5 |
Hb_027242_010 |
0.1351954997 |
- |
- |
PREDICTED: pathogenesis-related protein 1-like [Populus euphratica] |
| 6 |
Hb_002081_220 |
0.1384772079 |
- |
- |
PREDICTED: uncharacterized protein LOC103707552 [Phoenix dactylifera] |
| 7 |
Hb_006573_260 |
0.1389862244 |
- |
- |
- |
| 8 |
Hb_001102_060 |
0.1461509215 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At1g78530 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001101_020 |
0.1466940531 |
- |
- |
hypothetical protein RCOM_0615680 [Ricinus communis] |
| 10 |
Hb_187211_010 |
0.1486252562 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 11 |
Hb_000625_130 |
0.1531167126 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase [Jatropha curcas] |
| 12 |
Hb_000705_090 |
0.1536627653 |
- |
- |
PREDICTED: uncharacterized protein LOC100246830 [Vitis vinifera] |
| 13 |
Hb_028927_010 |
0.1553345978 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 14 |
Hb_106321_010 |
0.1562140597 |
- |
- |
PREDICTED: probable glutathione S-transferase [Jatropha curcas] |
| 15 |
Hb_010053_010 |
0.1583751871 |
- |
- |
hypothetical protein POPTR_0515s00220g [Populus trichocarpa] |
| 16 |
Hb_000566_110 |
0.1589416993 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Prunus mume] |
| 17 |
Hb_000075_030 |
0.1593305429 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C10-like [Fragaria vesca subsp. vesca] |
| 18 |
Hb_000832_250 |
0.1615191043 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC-like [Jatropha curcas] |
| 19 |
Hb_116469_010 |
0.1639075062 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |
| 20 |
Hb_000057_210 |
0.1644195733 |
- |
- |
hypothetical protein JCGZ_06274 [Jatropha curcas] |