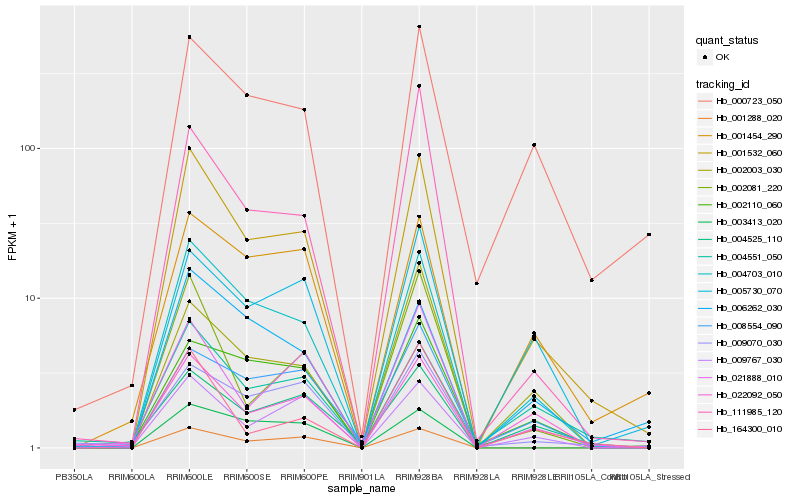
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001532_060 |
0.0 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 2 |
Hb_004703_010 |
0.0895790521 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 3 |
Hb_002081_220 |
0.1383126371 |
- |
- |
PREDICTED: uncharacterized protein LOC103707552 [Phoenix dactylifera] |
| 4 |
Hb_001288_020 |
0.1409161968 |
- |
- |
- |
| 5 |
Hb_021888_010 |
0.1480425512 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000723_050 |
0.1521480931 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 7 |
Hb_009767_030 |
0.1553548974 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 8 |
Hb_004551_050 |
0.1592863917 |
- |
- |
PREDICTED: glucose-6-phosphate/phosphate translocator 2, chloroplastic [Jatropha curcas] |
| 9 |
Hb_022092_050 |
0.1602243404 |
- |
- |
PREDICTED: uncharacterized protein LOC105136214 [Populus euphratica] |
| 10 |
Hb_009070_030 |
0.1611298471 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10 [Jatropha curcas] |
| 11 |
Hb_164300_010 |
0.16152973 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 12 |
Hb_006262_030 |
0.1619273694 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_005730_070 |
0.164181833 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 8 [Jatropha curcas] |
| 14 |
Hb_001454_290 |
0.1646507485 |
- |
- |
PREDICTED: uncharacterized protein LOC105643442 [Jatropha curcas] |
| 15 |
Hb_008554_090 |
0.1654032489 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_002003_030 |
0.1657436867 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 17 |
Hb_002110_060 |
0.1664101409 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 18 |
Hb_111985_120 |
0.168227276 |
- |
- |
dihydroflavonol-4-reductase [Vitis rotundifolia] |
| 19 |
Hb_004525_110 |
0.1756173025 |
- |
- |
hypothetical protein POPTR_0007s12850g [Populus trichocarpa] |
| 20 |
Hb_003413_020 |
0.1767590167 |
- |
- |
- |