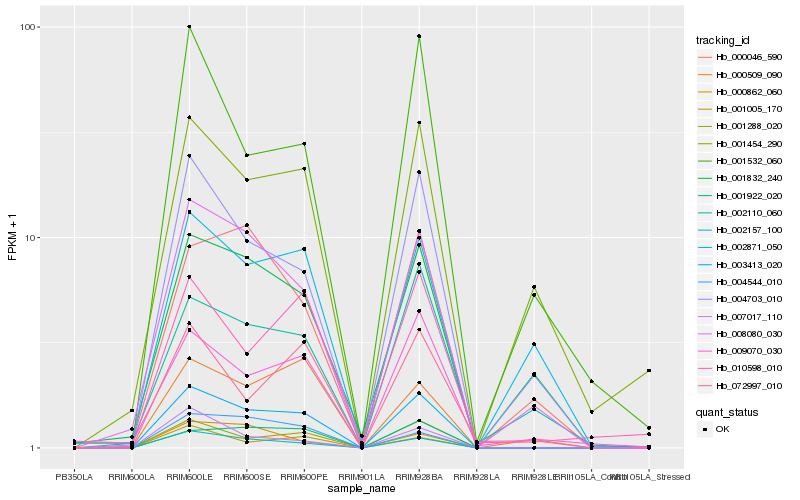
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003413_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_001288_020 |
0.0951397001 |
- |
- |
- |
| 3 |
Hb_002157_100 |
0.1005894663 |
- |
- |
- |
| 4 |
Hb_008080_030 |
0.1347511607 |
- |
- |
hypothetical protein POPTR_0275s002101g, partial [Populus trichocarpa] |
| 5 |
Hb_004703_010 |
0.1435939498 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 6 |
Hb_001922_020 |
0.1450899931 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001005_170 |
0.15242199 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 8 |
Hb_009070_030 |
0.1644357474 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10 [Jatropha curcas] |
| 9 |
Hb_001832_240 |
0.1724047236 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 10 |
Hb_002110_060 |
0.1757677626 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 11 |
Hb_010598_010 |
0.1758254922 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 12 |
Hb_001532_060 |
0.1767590167 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_004544_010 |
0.1770595069 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Jatropha curcas] |
| 14 |
Hb_072997_010 |
0.1796144238 |
- |
- |
hypothetical protein JCGZ_18743 [Jatropha curcas] |
| 15 |
Hb_000509_090 |
0.1808625245 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 16 |
Hb_001454_290 |
0.1813873217 |
- |
- |
PREDICTED: uncharacterized protein LOC105643442 [Jatropha curcas] |
| 17 |
Hb_007017_110 |
0.1837234899 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 18 |
Hb_000862_060 |
0.1851468282 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002871_050 |
0.1864372771 |
transcription factor |
TF Family: ARF |
auxin response factor 1 family protein [Populus trichocarpa] |
| 20 |
Hb_000046_590 |
0.1868963261 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |