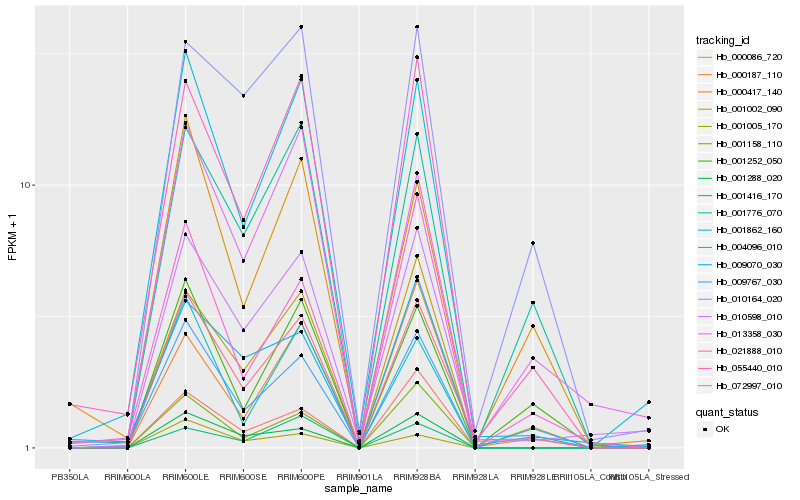
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_072997_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_18743 [Jatropha curcas] |
| 2 |
Hb_055440_010 |
0.1068469903 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_004096_010 |
0.1116089314 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 40 [Jatropha curcas] |
| 4 |
Hb_001002_090 |
0.1132300814 |
- |
- |
PREDICTED: receptor protein kinase-like protein At4g34220 [Jatropha curcas] |
| 5 |
Hb_010598_010 |
0.11615715 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 6 |
Hb_009767_030 |
0.121220673 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 7 |
Hb_001288_020 |
0.1276499395 |
- |
- |
- |
| 8 |
Hb_001158_110 |
0.1425828236 |
- |
- |
PREDICTED: uncharacterized protein LOC105648901 [Jatropha curcas] |
| 9 |
Hb_001776_070 |
0.1438045825 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase isoform X2 [Jatropha curcas] |
| 10 |
Hb_001252_050 |
0.149688252 |
- |
- |
Wall-associated receptor kinase-like 20 [Morus notabilis] |
| 11 |
Hb_001862_160 |
0.150027866 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.1 [Jatropha curcas] |
| 12 |
Hb_009070_030 |
0.1529133371 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10 [Jatropha curcas] |
| 13 |
Hb_010164_020 |
0.1566709434 |
- |
- |
PREDICTED: abscisate beta-glucosyltransferase-like [Jatropha curcas] |
| 14 |
Hb_013358_030 |
0.157028281 |
- |
- |
PREDICTED: U-box domain-containing protein 12-like [Jatropha curcas] |
| 15 |
Hb_000187_110 |
0.1583736807 |
- |
- |
PREDICTED: serine carboxypeptidase-like 7 [Jatropha curcas] |
| 16 |
Hb_021888_010 |
0.1609563183 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001005_170 |
0.1633765079 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 18 |
Hb_001416_170 |
0.1652879835 |
- |
- |
kinase, putative [Ricinus communis] |
| 19 |
Hb_000086_720 |
0.1674527965 |
- |
- |
leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 20 |
Hb_000417_140 |
0.1702003402 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 2 [Jatropha curcas] |