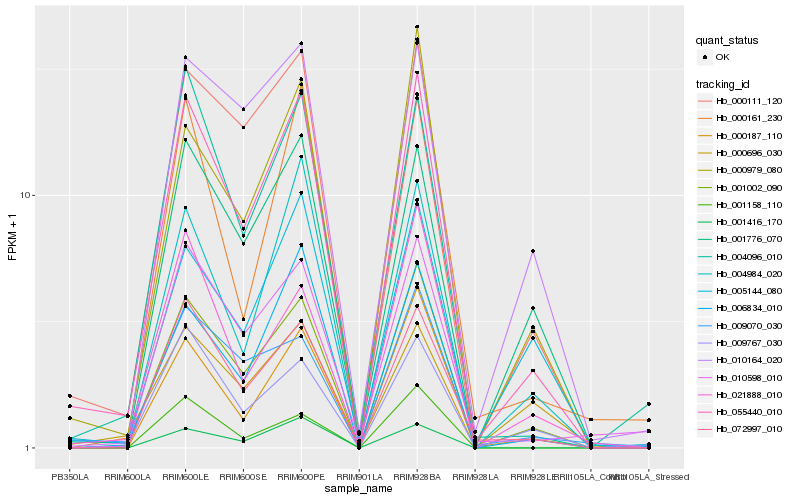
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_055440_010 |
0.0 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_001002_090 |
0.0739593003 |
- |
- |
PREDICTED: receptor protein kinase-like protein At4g34220 [Jatropha curcas] |
| 3 |
Hb_072997_010 |
0.1068469903 |
- |
- |
hypothetical protein JCGZ_18743 [Jatropha curcas] |
| 4 |
Hb_004984_020 |
0.1175253502 |
- |
- |
PREDICTED: uncharacterized protein LOC105647814 [Jatropha curcas] |
| 5 |
Hb_006834_010 |
0.1223517708 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004096_010 |
0.1224574326 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 40 [Jatropha curcas] |
| 7 |
Hb_000979_080 |
0.1245582227 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |
| 8 |
Hb_001776_070 |
0.1263769949 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase isoform X2 [Jatropha curcas] |
| 9 |
Hb_000187_110 |
0.1282161733 |
- |
- |
PREDICTED: serine carboxypeptidase-like 7 [Jatropha curcas] |
| 10 |
Hb_009070_030 |
0.1311307814 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10 [Jatropha curcas] |
| 11 |
Hb_009767_030 |
0.1326049378 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 12 |
Hb_010598_010 |
0.1339817644 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 13 |
Hb_001416_170 |
0.1394883624 |
- |
- |
kinase, putative [Ricinus communis] |
| 14 |
Hb_021888_010 |
0.1432614368 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000161_230 |
0.1478657794 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 16 |
Hb_010164_020 |
0.1491758333 |
- |
- |
PREDICTED: abscisate beta-glucosyltransferase-like [Jatropha curcas] |
| 17 |
Hb_001158_110 |
0.1513077072 |
- |
- |
PREDICTED: uncharacterized protein LOC105648901 [Jatropha curcas] |
| 18 |
Hb_005144_080 |
0.1540419101 |
- |
- |
Serine carboxypeptidase, putative [Ricinus communis] |
| 19 |
Hb_000696_030 |
0.1559857566 |
- |
- |
PREDICTED: uncharacterized protein LOC105647085 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000111_120 |
0.1574195425 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |