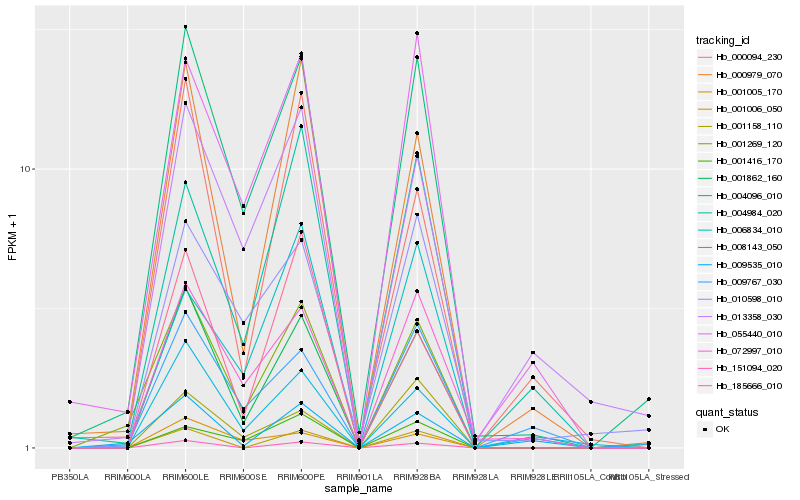
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001269_120 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF003-like [Jatropha curcas] |
| 2 |
Hb_004096_010 |
0.122228076 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 40 [Jatropha curcas] |
| 3 |
Hb_001862_160 |
0.1249124468 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.1 [Jatropha curcas] |
| 4 |
Hb_000979_070 |
0.150150255 |
- |
- |
PREDICTED: endoglucanase 8-like [Jatropha curcas] |
| 5 |
Hb_055440_010 |
0.1676487516 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_072997_010 |
0.1738457994 |
- |
- |
hypothetical protein JCGZ_18743 [Jatropha curcas] |
| 7 |
Hb_009535_010 |
0.1750805758 |
- |
- |
PREDICTED: DNA replication licensing factor MCM4 [Jatropha curcas] |
| 8 |
Hb_001416_170 |
0.1766409398 |
- |
- |
kinase, putative [Ricinus communis] |
| 9 |
Hb_001005_170 |
0.1845229358 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 10 |
Hb_185666_010 |
0.18609179 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3 [Jatropha curcas] |
| 11 |
Hb_009767_030 |
0.1912506481 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 12 |
Hb_000094_230 |
0.1915018781 |
- |
- |
hypothetical protein JCGZ_04416 [Jatropha curcas] |
| 13 |
Hb_013358_030 |
0.1920328352 |
- |
- |
PREDICTED: U-box domain-containing protein 12-like [Jatropha curcas] |
| 14 |
Hb_151094_020 |
0.1945237462 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Prunus mume] |
| 15 |
Hb_006834_010 |
0.1952453971 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004984_020 |
0.195339893 |
- |
- |
PREDICTED: uncharacterized protein LOC105647814 [Jatropha curcas] |
| 17 |
Hb_010598_010 |
0.1965717879 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 18 |
Hb_008143_050 |
0.1969403025 |
- |
- |
PREDICTED: cytochrome P450 710A1-like [Jatropha curcas] |
| 19 |
Hb_001006_050 |
0.1981414369 |
- |
- |
PREDICTED: uncharacterized protein LOC101777526 [Setaria italica] |
| 20 |
Hb_001158_110 |
0.1984140139 |
- |
- |
PREDICTED: uncharacterized protein LOC105648901 [Jatropha curcas] |