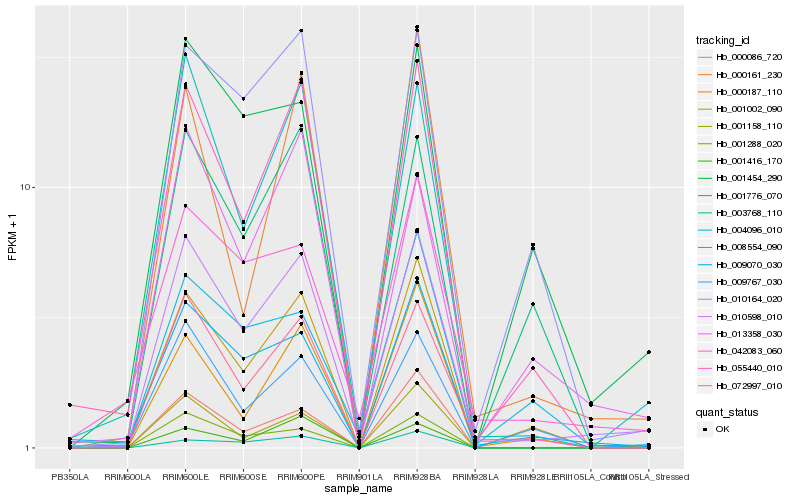
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010598_010 |
0.0 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 2 |
Hb_004096_010 |
0.1074386192 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 40 [Jatropha curcas] |
| 3 |
Hb_072997_010 |
0.11615715 |
- |
- |
hypothetical protein JCGZ_18743 [Jatropha curcas] |
| 4 |
Hb_009070_030 |
0.1297396018 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10 [Jatropha curcas] |
| 5 |
Hb_055440_010 |
0.1339817644 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_001002_090 |
0.1341515222 |
- |
- |
PREDICTED: receptor protein kinase-like protein At4g34220 [Jatropha curcas] |
| 7 |
Hb_001288_020 |
0.1401867351 |
- |
- |
- |
| 8 |
Hb_042083_060 |
0.1475068858 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g45780 [Jatropha curcas] |
| 9 |
Hb_000086_720 |
0.1476533833 |
- |
- |
leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 10 |
Hb_009767_030 |
0.1478350537 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 11 |
Hb_013358_030 |
0.1480191961 |
- |
- |
PREDICTED: U-box domain-containing protein 12-like [Jatropha curcas] |
| 12 |
Hb_010164_020 |
0.1555093378 |
- |
- |
PREDICTED: abscisate beta-glucosyltransferase-like [Jatropha curcas] |
| 13 |
Hb_001158_110 |
0.1628855879 |
- |
- |
PREDICTED: uncharacterized protein LOC105648901 [Jatropha curcas] |
| 14 |
Hb_001454_290 |
0.166187442 |
- |
- |
PREDICTED: uncharacterized protein LOC105643442 [Jatropha curcas] |
| 15 |
Hb_001776_070 |
0.1688838537 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase isoform X2 [Jatropha curcas] |
| 16 |
Hb_000187_110 |
0.1693843518 |
- |
- |
PREDICTED: serine carboxypeptidase-like 7 [Jatropha curcas] |
| 17 |
Hb_000161_230 |
0.1694003081 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 18 |
Hb_008554_090 |
0.1699788418 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_003768_110 |
0.1722508179 |
- |
- |
hypothetical protein POPTR_0004s12230g [Populus trichocarpa] |
| 20 |
Hb_001416_170 |
0.1730963506 |
- |
- |
kinase, putative [Ricinus communis] |