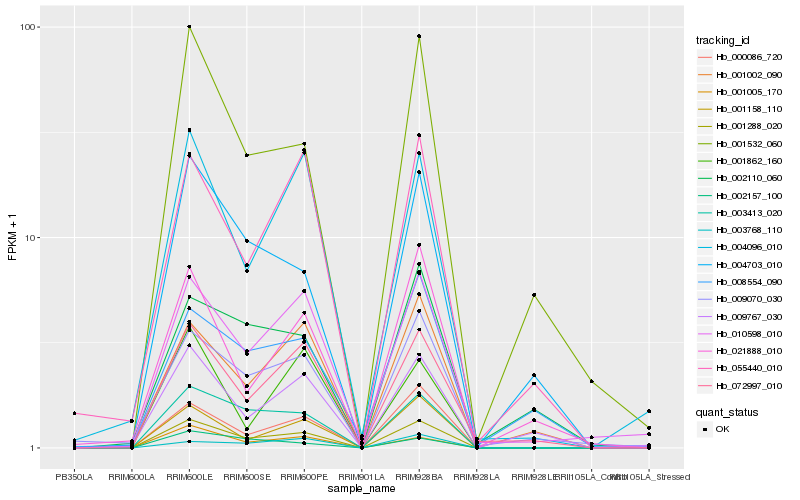
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001288_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_003413_020 |
0.0951397001 |
- |
- |
- |
| 3 |
Hb_001158_110 |
0.1099854449 |
- |
- |
PREDICTED: uncharacterized protein LOC105648901 [Jatropha curcas] |
| 4 |
Hb_000086_720 |
0.123356017 |
- |
- |
leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 5 |
Hb_072997_010 |
0.1276499395 |
- |
- |
hypothetical protein JCGZ_18743 [Jatropha curcas] |
| 6 |
Hb_001005_170 |
0.1398013969 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 7 |
Hb_010598_010 |
0.1401867351 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 8 |
Hb_001532_060 |
0.1409161968 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 9 |
Hb_004096_010 |
0.1445392724 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 40 [Jatropha curcas] |
| 10 |
Hb_009070_030 |
0.1460883719 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10 [Jatropha curcas] |
| 11 |
Hb_004703_010 |
0.1490720633 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 12 |
Hb_001002_090 |
0.1491038135 |
- |
- |
PREDICTED: receptor protein kinase-like protein At4g34220 [Jatropha curcas] |
| 13 |
Hb_009767_030 |
0.1606612219 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 14 |
Hb_055440_010 |
0.162282275 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_002157_100 |
0.1681226611 |
- |
- |
- |
| 16 |
Hb_021888_010 |
0.1688196528 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003768_110 |
0.1772629227 |
- |
- |
hypothetical protein POPTR_0004s12230g [Populus trichocarpa] |
| 18 |
Hb_001862_160 |
0.1816781886 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.1 [Jatropha curcas] |
| 19 |
Hb_002110_060 |
0.1853887094 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 20 |
Hb_008554_090 |
0.1875802892 |
- |
- |
zinc finger protein, putative [Ricinus communis] |