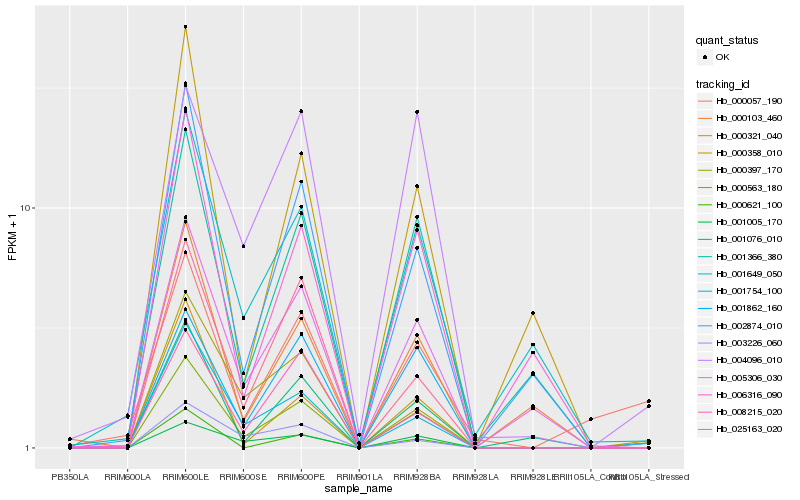
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000563_180 |
0.0 |
- |
- |
PREDICTED: J protein JJJ2 [Jatropha curcas] |
| 2 |
Hb_001754_100 |
0.1464345235 |
- |
- |
PREDICTED: peroxidase 5-like [Jatropha curcas] |
| 3 |
Hb_001005_170 |
0.1547018479 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 4 |
Hb_002874_010 |
0.1624009889 |
- |
- |
PREDICTED: replication protein A 32 kDa subunit A isoform X1 [Jatropha curcas] |
| 5 |
Hb_005306_030 |
0.1630005971 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 [Jatropha curcas] |
| 6 |
Hb_001862_160 |
0.1630385312 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.1 [Jatropha curcas] |
| 7 |
Hb_001366_380 |
0.167393719 |
- |
- |
PREDICTED: pectin acetylesterase 6-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_025163_020 |
0.1704619017 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 9 |
Hb_008215_020 |
0.1727333055 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_001076_010 |
0.1759914846 |
transcription factor |
TF Family: B3 |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_000397_170 |
0.1772775913 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000057_190 |
0.1798831746 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001649_050 |
0.1817970079 |
- |
- |
Shaggy-like kinase 13 isoform 1 [Theobroma cacao] |
| 14 |
Hb_000321_040 |
0.1830061789 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 5 [Jatropha curcas] |
| 15 |
Hb_003226_060 |
0.1877171827 |
- |
- |
Pectinesterase-2 precursor, putative [Ricinus communis] |
| 16 |
Hb_006316_090 |
0.1916758807 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 17 |
Hb_004096_010 |
0.1918269171 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 40 [Jatropha curcas] |
| 18 |
Hb_000103_460 |
0.1922056562 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 19 |
Hb_000358_010 |
0.1932403182 |
- |
- |
PREDICTED: basic blue protein [Populus euphratica] |
| 20 |
Hb_000621_100 |
0.1935301518 |
- |
- |
conserved hypothetical protein [Ricinus communis] |